Genomic and proteomic analyses of the fungus Arthrobotrys oligospora provide insights into nematode-trap formation
- PMID: 21909256
- PMCID: PMC3164635
- DOI: 10.1371/journal.ppat.1002179
Genomic and proteomic analyses of the fungus Arthrobotrys oligospora provide insights into nematode-trap formation
Abstract
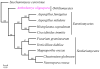
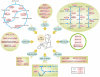
Nematode-trapping fungi are "carnivorous" and attack their hosts using specialized trapping devices. The morphological development of these traps is the key indicator of their switch from saprophytic to predacious lifestyles. Here, the genome of the nematode-trapping fungus Arthrobotrys oligospora Fres. (ATCC24927) was reported. The genome contains 40.07 Mb assembled sequence with 11,479 predicted genes. Comparative analysis showed that A. oligospora shared many more genes with pathogenic fungi than with non-pathogenic fungi. Specifically, compared to several sequenced ascomycete fungi, the A. oligospora genome has a larger number of pathogenicity-related genes in the subtilisin, cellulase, cellobiohydrolase, and pectinesterase gene families. Searching against the pathogen-host interaction gene database identified 398 homologous genes involved in pathogenicity in other fungi. The analysis of repetitive sequences provided evidence for repeat-induced point mutations in A. oligospora. Proteomic and quantitative PCR (qPCR) analyses revealed that 90 genes were significantly up-regulated at the early stage of trap-formation by nematode extracts and most of these genes were involved in translation, amino acid metabolism, carbohydrate metabolism, cell wall and membrane biogenesis. Based on the combined genomic, proteomic and qPCR data, a model for the formation of nematode trapping device in this fungus was proposed. In this model, multiple fungal signal transduction pathways are activated by its nematode prey to further regulate downstream genes associated with diverse cellular processes such as energy metabolism, biosynthesis of the cell wall and adhesive proteins, cell division, glycerol accumulation and peroxisome biogenesis. This study will facilitate the identification of pathogenicity-related genes and provide a broad foundation for understanding the molecular and evolutionary mechanisms underlying fungi-nematodes interactions.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

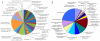



Comment in
-
Fungal pathogenesis: hungry fungus eats nematode.Nat Rev Microbiol. 2011 Oct 3;9(11):766-7. doi: 10.1038/nrmicro2674. Nat Rev Microbiol. 2011. PMID: 21963804 No abstract available.
References
-
- Pramer D. Nematode-trapping fungi. Science. 1964;144:382–388. - PubMed
-
- Nordbring-Hertz B, Jansson HB, Tunlid A. Chichester: John Wiley & Sons, Ltd; 2006. Nematophagous fungi. In Encyclopedia of life sciences.
-
- Schmidt AR, Dörfelt H, Perrichot V. Carnivorous fungi from Cretaceous Amber. Science. 2007;318:1743. - PubMed
-
- Nordbring-Hertz B. Morphogenesis in the nematode-trapping fungus Arthrobotrys oligospora-an extensive plasticity of infection structures. Mycologist. 2004;18:125–133.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

