c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress
- PMID: 21909268
- PMCID: PMC3164647
- DOI: 10.1371/journal.ppat.1002217
c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress
Abstract
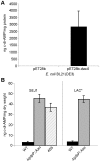
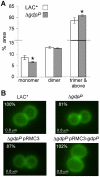
The cell wall is a vital and multi-functional part of bacterial cells. For Staphylococcus aureus, an important human bacterial pathogen, surface proteins and cell wall polymers are essential for adhesion, colonization and during the infection process. One such cell wall polymer, lipoteichoic acid (LTA), is crucial for normal bacterial growth and cell division. Upon depletion of this polymer bacteria increase in size and a misplacement of division septa and eventual cell lysis is observed. In this work, we describe the isolation and characterization of LTA-deficient S. aureus suppressor strains that regained the ability to grow almost normally in the absence of this cell wall polymer. Using a whole genome sequencing approach, compensatory mutations were identified and revealed that mutations within one gene, gdpP (GGDEF domain protein containing phosphodiesterase), allow both laboratory and clinical isolates of S. aureus to grow without LTA. It was determined that GdpP has phosphodiesterase activity in vitro and uses the cyclic dinucleotide c-di-AMP as a substrate. Furthermore, we show for the first time that c-di-AMP is produced in S. aureus presumably by the S. aureus DacA protein, which has diadenylate cyclase activity. We also demonstrate that GdpP functions in vivo as a c-di-AMP-specific phosphodiesterase, as intracellular c-di-AMP levels increase drastically in gdpP deletion strains and in an LTA-deficient suppressor strain. An increased amount of cross-linked peptidoglycan was observed in the gdpP mutant strain, a cell wall alteration that could help bacteria compensate for the lack of LTA. Lastly, microscopic analysis of wild-type and gdpP mutant strains revealed a 13-22% reduction in the cell size of bacteria with increased c-di-AMP levels. Taken together, these data suggest a function for this novel secondary messenger in controlling cell size of S. aureus and in helping bacteria to cope with extreme membrane and cell wall stress.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Francis JS, Doherty MC, Lopatin U, Johnston CP, Sinha G, et al. Severe community-onset pneumonia in healthy adults caused by methicillin-resistant Staphylococcus aureus carrying the Panton-Valentine leukocidin genes. Clin Infect Dis. 2005;40:100–1007. - PubMed
-
- Fridkin SK, Hageman JC, Morrison M, Sanza LT, Como-Sabetti K, et al. Methicillin-resistant Staphylococcus aureus disease in three communities. N Engl J Med. 2005;352:1436–1444. - PubMed
-
- Bhavsar AP, Brown ED. Cell wall assembly in Bacillus subtilis: how spirals and spaces challenge paradigms. Mol Microbiol. 2006;60:1077–1090. - PubMed
-
- Xia G, Kohler T, Peschel A. The wall teichoic acid and lipoteichoic acid polymers of Staphylococcus aureus. Int J Med Microbiol. 2010;300:148–154. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

