No evidence for strong recent positive selection favoring the 7 repeat allele of VNTR in the DRD4 gene
- PMID: 21909391
- PMCID: PMC3164202
- DOI: 10.1371/journal.pone.0024410
No evidence for strong recent positive selection favoring the 7 repeat allele of VNTR in the DRD4 gene
Abstract
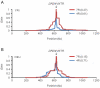
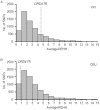
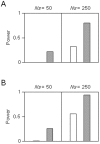
The human dopamine receptor D4 (DRD4) gene contains a 48-bp variable number of tandem repeat (VNTR) in exon 3, encoding the third intracellular loop of this dopamine receptor. The DRD4 7R allele, which seems to have a single origin, is commonly observed in various human populations and the nucleotide diversity of the DRD4 7R haplotype at the DRD4 locus is reduced compared to the most common DRD4 4R haplotype. Based on these observations, previous studies have hypothesized that positive selection has acted on the DRD4 7R allele. However, the degrees of linkage disequilibrium (LD) of the DRD4 7R allele with single nucleotide polymorphisms (SNPs) outside the DRD4 locus have not been evaluated. In this study, to re-examine the possibility of recent positive selection favoring the DRD4 7R allele, we genotyped HapMap subjects for DRD4 VNTR, and conducted several neutrality tests including long range haplotype test and iHS test based on the extended haplotype homozygosity. Our results indicated that LD of the DRD4 7R allele was not extended compared to SNP alleles with the similar frequency. Thus, we conclude that the DRD4 7R allele has not been subjected to strong recent positive selection.
Conflict of interest statement
Figures



Similar articles
-
The genetic architecture of selection at the human dopamine receptor D4 (DRD4) gene locus.Am J Hum Genet. 2004 May;74(5):931-44. doi: 10.1086/420854. Epub 2004 Apr 9. Am J Hum Genet. 2004. PMID: 15077199 Free PMC article.
-
Increased brain activity to unpleasant stimuli in individuals with the 7R allele of the DRD4 gene.Psychiatry Res. 2015 Jan 30;231(1):58-63. doi: 10.1016/j.pscychresns.2014.10.021. Epub 2014 Nov 4. Psychiatry Res. 2015. PMID: 25481571 Free PMC article.
-
Novelty seeking and the dopamine D4 receptor gene (DRD4) revisited in Asians: haplotype characterization and relevance of the 2-repeat allele.Am J Med Genet B Neuropsychiatr Genet. 2007 Jun 5;144B(4):453-7. doi: 10.1002/ajmg.b.30473. Am J Med Genet B Neuropsychiatr Genet. 2007. PMID: 17474081
-
Underlying Susceptibility to Eating Disorders and Drug Abuse: Genetic and Pharmacological Aspects of Dopamine D4 Receptors.Nutrients. 2020 Jul 30;12(8):2288. doi: 10.3390/nu12082288. Nutrients. 2020. PMID: 32751662 Free PMC article. Review.
-
Parsing out the role of dopamine D4 receptor gene (DRD4) on alcohol-related phenotypes: A meta-analysis and systematic review.Addict Biol. 2020 May;25(3):e12770. doi: 10.1111/adb.12770. Epub 2019 May 31. Addict Biol. 2020. PMID: 31149768 Free PMC article.
Cited by
-
The Attention-Deficit/Hyperactivity Disorder-Associated DRD4 7R Allele Predicts Household Economic Status but Not Nutritional Status in Northern Kenyan Rendille Children.Am J Hum Biol. 2025 Mar;37(3):e70027. doi: 10.1002/ajhb.70027. Am J Hum Biol. 2025. PMID: 40062537
-
Humanized dopamine D4.7 receptor male mice display risk-taking behavior and deficits of social recognition and working memory in light/dark-dependent manner.J Neurosci Res. 2024 Feb;102(2):e25299. doi: 10.1002/jnr.25299. J Neurosci Res. 2024. PMID: 38361407 Free PMC article. Review.
-
Prevalence of Common Alleles of Some Stress Resilience Genes among Adolescents Born in Different Periods Relative to the Socioeconomic Crisis of the 1990s in Russia.Curr Issues Mol Biol. 2022 Dec 21;45(1):51-65. doi: 10.3390/cimb45010004. Curr Issues Mol Biol. 2022. PMID: 36661490 Free PMC article.
-
The dopamine hypothesis for ADHD: An evaluation of evidence accumulated from human studies and animal models.Front Psychiatry. 2024 Nov 15;15:1492126. doi: 10.3389/fpsyt.2024.1492126. eCollection 2024. Front Psychiatry. 2024. PMID: 39619336 Free PMC article. Review.
-
Signatures of positive selection in the cis-regulatory sequences of the human oxytocin receptor (OXTR) and arginine vasopressin receptor 1a (AVPR1A) genes.BMC Evol Biol. 2015 May 13;15:85. doi: 10.1186/s12862-015-0372-7. BMC Evol Biol. 2015. PMID: 25968600 Free PMC article.
References
-
- Van Tol HH, Wu CM, Guan HC, Ohara K, Bunzow JR, et al. Multiple dopamine D4 receptor variants in the human population. Nature. 1992;358:149–152. - PubMed
-
- Chang FM, Kidd JR, Livak KJ, Pakstis AJ, Kidd KK. The world-wide distribution of allele frequencies at the human dopamine D4 receptor locus. Hum Genet. 1996;98:91–101. - PubMed
-
- Asghari V, Sanyal S, Buchwaldt S, Paterson A, Jovanovic V, et al. Modulation of intracellular cyclic AMP levels by different human dopamine D4 receptor variants. J Neurochem. 1995;65:1157–1165. - PubMed
-
- Schoots O, Van Tol HH. The human dopamine D4 receptor repeat sequences modulate expression. Pharmacogenomics J. 2003;3:343–348. - PubMed
-
- Ebstein RP, Novick O, Umansky R, Priel B, Osher Y, et al. Dopamine D4 receptor (D4DR) exon III polymorphism associated with the human personality trait of Novelty Seeking. Nat Genet. 1996;12:78–80. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

