Deciphering network community structure by surprise
- PMID: 21909420
- PMCID: PMC3164713
- DOI: 10.1371/journal.pone.0024195
Deciphering network community structure by surprise
Abstract
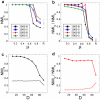
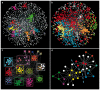
The analysis of complex networks permeates all sciences, from biology to sociology. A fundamental, unsolved problem is how to characterize the community structure of a network. Here, using both standard and novel benchmarks, we show that maximization of a simple global parameter, which we call Surprise (S), leads to a very efficient characterization of the community structure of complex synthetic networks. Particularly, S qualitatively outperforms the most commonly used criterion to define communities, Newman and Girvan's modularity (Q). Applying S maximization to real networks often provides natural, well-supported partitions, but also sometimes counterintuitive solutions that expose the limitations of our previous knowledge. These results indicate that it is possible to define an effective global criterion for community structure and open new routes for the understanding of complex networks.
Conflict of interest statement
Figures



References
-
- Barabási AL, Oltvai ZN. Network biology: Understanding the cell's functional organization. Nat Rev Genet. 2004;5:101–113. - PubMed
-
- Wasserman S, Faust K. Social network analysis: Methods and applications. 1994. Cambridge University Press, Cambridge, U.K.
-
- Strogatz SH. Exploring complex networks. Nature. 2001;410:268–276. - PubMed
-
- Costa LD, Rodrigues FA, Travieso G, Boas PRV. Characterization of complex networks: A survey of measurements. Adv Phys. 2007;56:167–242.
-
- Newman MEJ. Networks: An introduction. 2010. Oxford University Press, Oxford. U.K.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

