Cap-independent translation promotes C. elegans germ cell apoptosis through Apaf-1/CED-4 in a caspase-dependent mechanism
- PMID: 21909434
- PMCID: PMC3164730
- DOI: 10.1371/journal.pone.0024444
Cap-independent translation promotes C. elegans germ cell apoptosis through Apaf-1/CED-4 in a caspase-dependent mechanism
Abstract
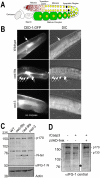
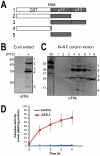
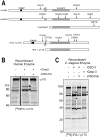
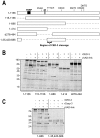
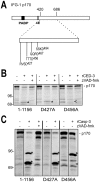
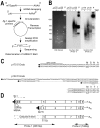
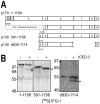
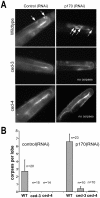
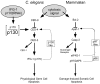
Apoptosis is a natural process during animal development for the programmed removal of superfluous cells. During apoptosis general protein synthesis is reduced, but the synthesis of cell death proteins is enhanced. Selective translation has been attributed to modification of the protein synthesis machinery to disrupt cap-dependent mRNA translation and induce a cap-independent mechanism. We have previously shown that disruption of the balance between cap-dependent and cap-independent C. elegans eIF4G isoforms (IFG-1 p170 and p130) by RNA interference promotes apoptosis in developing oocytes. Germ cell apoptosis was accompanied by the appearance of the Apaf-1 homolog, CED-4. Here we show that IFG-1 p170 is a native substrate of the worm executioner caspase, CED-3, just as mammalian eIF4GI is cleaved by caspase-3. Loss of Bcl-2 function (ced-9ts) in worms induced p170 cleavage in vivo, coincident with extensive germ cell apoptosis. Truncation of IFG-1 occurred at a single site that separates the cap-binding and ribosome-associated domains. Site-directed mutagenesis indicated that CED-3 processes IFG-1 at a non-canonical motif, TTTD(456). Coincidentally, the recognition site was located 65 amino acids downstream of the newly mapped IFG-1 p130 start site suggesting that both forms support cap-independent initiation. Genetic evidence confirmed that apoptosis induced by loss of ifg-1 p170 mRNA was caspase (ced-3) and apoptosome (ced-4/Apaf-1) dependent. These findings support a new paradigm in which modal changes in protein synthesis act as a physiological signal to initiate cell death, rather than occur merely as downstream consequences of the apoptotic event.
Conflict of interest statement
Figures
Similar articles
-
Depletion of the cap-associated isoform of translation factor eIF4G induces germline apoptosis in C. elegans.Cell Death Differ. 2008 Aug;15(8):1232-42. doi: 10.1038/cdd.2008.46. Epub 2008 May 2. Cell Death Differ. 2008. PMID: 18451872
-
Apoptosome assembly.Methods Enzymol. 2008;442:141-56. doi: 10.1016/S0076-6879(08)01407-9. Methods Enzymol. 2008. PMID: 18662568
-
Both the caspase CSP-1 and a caspase-independent pathway promote programmed cell death in parallel to the canonical pathway for apoptosis in Caenorhabditis elegans.PLoS Genet. 2013;9(3):e1003341. doi: 10.1371/journal.pgen.1003341. Epub 2013 Mar 7. PLoS Genet. 2013. PMID: 23505386 Free PMC article.
-
New insights into apoptosome structure and function.Cell Death Differ. 2018 Jul;25(7):1194-1208. doi: 10.1038/s41418-017-0025-z. Epub 2018 May 15. Cell Death Differ. 2018. PMID: 29765111 Free PMC article. Review.
-
Apoptosomes: engines for caspase activation.Curr Opin Cell Biol. 2002 Dec;14(6):715-20. doi: 10.1016/s0955-0674(02)00381-2. Curr Opin Cell Biol. 2002. PMID: 12473344 Review.
Cited by
-
Reducing translation through eIF4G/IFG-1 improves survival under ER stress that depends on heat shock factor HSF-1 in Caenorhabditis elegans.Aging Cell. 2016 Dec;15(6):1027-1038. doi: 10.1111/acel.12516. Epub 2016 Aug 18. Aging Cell. 2016. PMID: 27538368 Free PMC article.
-
Functional Interactions Between rsks-1/S6K, glp-1/Notch, and Regulators of Caenorhabditis elegans Fertility and Germline Stem Cell Maintenance.G3 (Bethesda). 2018 Oct 3;8(10):3293-3309. doi: 10.1534/g3.118.200511. G3 (Bethesda). 2018. PMID: 30126834 Free PMC article.
-
RNA profiles of porcine embryos during genome activation reveal complex metabolic switch sensitive to in vitro conditions.PLoS One. 2013 Apr 29;8(4):e61547. doi: 10.1371/journal.pone.0061547. Print 2013. PLoS One. 2013. PMID: 23637850 Free PMC article.
-
Role of translation initiation factor 4G in lifespan regulation and age-related health.Ageing Res Rev. 2014 Jan;13:115-24. doi: 10.1016/j.arr.2013.12.008. Epub 2014 Jan 3. Ageing Res Rev. 2014. PMID: 24394551 Free PMC article. Review.
-
Cap-Independent mRNA Translation in Germ Cells.Int J Mol Sci. 2019 Jan 5;20(1):173. doi: 10.3390/ijms20010173. Int J Mol Sci. 2019. PMID: 30621249 Free PMC article. Review.
References
-
- Graber TE, Holcik M. Cap-independent regulation of gene expression in apoptosis. Mol Biosyst. 2007;3:825–834. - PubMed
-
- Keiper BD, Gan W, Rhoads RE. Protein synthesis initiation factor 4G. Int J Biochem Cell Biol. 1999;31:37–41. - PubMed
-
- Keiper BD, Rhoads RE. Translational recruitment of Xenopus maternal mRNAs in response to poly(A) elongation requires initiation factor eIF4G-1. Dev Biol. 1999;206:1–14. - PubMed
-
- Lazebnik YA, Kaufmann SH, Desnoyers S, Poirier GG, Earnshaw WC. Cleavage of poly(ADP-ribose) polymerase by a proteinase with properties like ICE. Nature. 1994;371:346–347. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

