Prediction of the pathogens that are the cause of pneumonia by the battlefield hypothesis
- PMID: 21909436
- PMCID: PMC3164732
- DOI: 10.1371/journal.pone.0024474
Prediction of the pathogens that are the cause of pneumonia by the battlefield hypothesis
Abstract
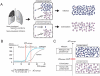
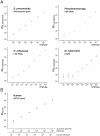
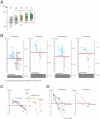
Commensal organisms are frequent causes of pneumonia. However, the detection of these organisms in the airway does not mean that they are the causative pathogens; they may exist merely as colonizers. In up to 50% cases of pneumonia, the causative pathogens remain unidentified, thereby hampering targeting therapies. In speculating on the role of a commensal organism in pneumonia, we devised the battlefield hypothesis. In the "pneumonia battlefield," the organism-to-human cell number ratio may be an index for the pathogenic role of the organism. Using real-time PCR reactions for sputum samples, we tested whether the hypothesis predicts the results of bacteriological clinical tests for 4 representative commensal organisms: Streptococcus pneumoniae, Haemophilus influenzae, Pseudomonas spp., and Moraxella catarrhalis. The cutoff value for the organism-to-human cell number ratio, above which the pathogenic role of the organism was suspected, was set up for each organism using 224 sputum samples. The validity of the cutoff value was then tested in a prospective study that included 153 samples; the samples were classified into 3 groups, and each group contained 93%, 7%, and 0% of the samples from pneumonia, in which the pathogenic role of Streptococcus pneumoniae was suggested by the clinical tests. The results for Haemophilus influenzae, Pseudomonas spp., and Moraxella catarrhalis were 100%, 0%, and 0%, respectively. The battlefield hypothesis enabled legitimate interpretation of the PCR results and predicted pneumonia in which the pathogenic role of the organism was suggested by the clinical test. The PCR reactions based on the battlefield hypothesis may help to promote targeted therapies for pneumonia. The prospective observatory study described in the current report had been registered to the University Hospital Medical Information Network (UMIN) registry before its initiation, where the UMIN is a registry approved by the International Committee of Medical Journal Editors (ICMJE). The UMIN registry number was UMIN000001118: A prospective study for the investigation of the validity of cutoff values established for the HIRA-TAN system (April 9, 2008).
Conflict of interest statement
Figures

References
-
- Seifert HS, DiRita VJ. Evolution of Microbial Pathogens: Blackwell Publishing 2006.
-
- American Thoracic Society; Infectious Diseases Society of America. Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. Am J Respir Crit Care Med. 2005;171:388–416. - PubMed
-
- Mandell LA, Marrie TJ, Grossman RF, Chow AW, Hyland RH. Canadian guidelines for the initial management of community-acquired pneumonia: an evidence-based update by the Canadian Infectious Diseases Society and the Canadian Thoracic Society. The Canadian Community-Acquired Pneumonia Working Group. Clin Infect Dis. 2000;31:383–421. - PubMed
-
- Woodhead M, Blasi F, Ewig S, Huchon G, Ieven M, et al. Guidelines for the management of adult lower respiratory tract infections. Eur Respir J. 2005;26:1138–1180. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

