Dynamics and control of state-dependent networks for probing genomic organization
- PMID: 21911407
- PMCID: PMC3198315
- DOI: 10.1073/pnas.1113249108
Dynamics and control of state-dependent networks for probing genomic organization
Abstract
A state-dependent dynamic network is a collection of elements that interact through a network, whose geometry evolves as the state of the elements changes over time. The genome is an intriguing example of a state-dependent network, where chromosomal geometry directly relates to genomic activity, which in turn strongly correlates with geometry. Here we examine various aspects of a genomic state-dependent dynamic network. In particular, we elaborate on one of the important ramifications of viewing genomic networks as being state-dependent, namely, their controllability during processes of genomic reorganization such as in cell differentiation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




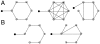
 and
and  are determined following an intricate alignment between form and function.
are determined following an intricate alignment between form and function.
 that steers the initial zero state at infinite past to a point on the unit ball at t = 0 (Top). Moreover, this Gramian defines how inputs on the unit sphere (Bottom Left), such as noise or external signals, map to system states: Directions that are more controllable are characterized by elongated ellipsoidal axes, whereas the shortened axes are less controllable directions (Bottom Right).
that steers the initial zero state at infinite past to a point on the unit ball at t = 0 (Top). Moreover, this Gramian defines how inputs on the unit sphere (Bottom Left), such as noise or external signals, map to system states: Directions that are more controllable are characterized by elongated ellipsoidal axes, whereas the shortened axes are less controllable directions (Bottom Right).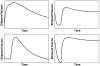

Comment in
-
Mathematical controllability of genomic networks.Proc Natl Acad Sci U S A. 2011 Oct 18;108(42):17243-4. doi: 10.1073/pnas.1114108108. Epub 2011 Oct 10. Proc Natl Acad Sci U S A. 2011. PMID: 21987806 Free PMC article. No abstract available.
References
-
- Strogatz SH. Exploring complex networks. Nature. 2001;410:268–276. - PubMed
-
- Diestel R. Graph Theory. Heidelberg, Germany: Springer; 2000.
-
- Cook P. Predicting three-dimensional genome structure from transcriptional activity. Nat Genet. 2002;32:347–352. - PubMed
-
- Weintraub H. Assembly and propagation of repressed and derepressed chromosomal states. Cell. 1985;42:705–711. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

