Tsx produces a long noncoding RNA and has general functions in the germline, stem cells, and brain
- PMID: 21912526
- PMCID: PMC3164691
- DOI: 10.1371/journal.pgen.1002248
Tsx produces a long noncoding RNA and has general functions in the germline, stem cells, and brain
Abstract
The Tsx gene resides at the X-inactivation center and is thought to encode a protein expressed in testis, but its function has remained mysterious. Given its proximity to noncoding genes that regulate X-inactivation, here we characterize Tsx and determine its function in mice. We find that Tsx is actually noncoding and the long transcript is expressed robustly in meiotic germ cells, embryonic stem cells, and brain. Targeted deletion of Tsx generates viable offspring and X-inactivation is only mildly affected in embryonic stem cells. However, mutant embryonic stem cells are severely growth-retarded, differentiate poorly, and show elevated cell death. Furthermore, male mice have smaller testes resulting from pachytene-specific apoptosis and a maternal-specific effect results in slightly smaller litters. Intriguingly, male mice lacking Tsx are less fearful and have measurably enhanced hippocampal short-term memory. Combined, our study indicates that Tsx performs general functions in multiple cell types and links the noncoding locus to stem and germ cell development, learning, and behavior in mammals.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
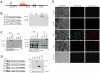




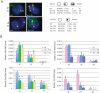

References
-
- Wutz A, Gribnau J. X inactivation Xplained. Curr Opin Genet Dev. 2007;17:387–393. - PubMed
-
- Lucchesi JC, Kelly WG, Panning B. Chromatin remodeling in dosage compensation. Annu Rev Genet. 2005;39:615–651. - PubMed
-
- Payer B, Lee JT. X chromosome dosage compensation: how mammals keep the balance. Annu Rev Genet. 2008;42:733–772. - PubMed
-
- Jonkers I, Barakat TS, Achame EM, Monkhorst K, Kenter A, et al. RNF12 is an X-Encoded dose-dependent activator of X chromosome inactivation. Cell. 2009;139:999–1011. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

