The mechanism of insulin-stimulated 4E-BP protein binding to mammalian target of rapamycin (mTOR) complex 1 and its contribution to mTOR complex 1 signaling
- PMID: 21914810
- PMCID: PMC3207408
- DOI: 10.1074/jbc.M111.245449
The mechanism of insulin-stimulated 4E-BP protein binding to mammalian target of rapamycin (mTOR) complex 1 and its contribution to mTOR complex 1 signaling
Abstract
Insulin activation of mTOR complex 1 is accompanied by enhanced binding of substrates. We examined the mechanism and contribution of this enhancement to insulin activation of mTORC1 signaling in 293E and HeLa cells. In 293E, insulin increased the amount of mTORC1 retrieved by the transiently expressed nonphosphorylatable 4E-BP[5A] to an extent that varied inversely with the amount of PRAS40 bound to mTORC1. RNAi depletion of PRAS40 enhanced 4E-BP[5A] binding to ∼70% the extent of maximal insulin, and PRAS40 RNAi and insulin together did not increase 4E-BP[5A] binding beyond insulin alone, suggesting that removal of PRAS40 from mTORC1 is the predominant mechanism of an insulin-induced increase in substrate access. As regards the role of increased substrate access in mTORC1 signaling, RNAi depletion of PRAS40, although increasing 4E-BP[5A] binding, did not stimulate phosphorylation of endogenous mTORC1 substrates S6K1(Thr(389)) or 4E-BP (Thr(37)/Thr(46)), the latter already ∼70% of maximal in amino acid replete, serum-deprived 293E cells. In HeLa cells, insulin and PRAS40 RNAi also both enhanced the binding of 4E-BP[5A] to raptor but only insulin stimulated S6K1 and 4E-BP phosphorylation. Furthermore, Rheb overexpression in 293E activated mTORC1 signaling completely without causing PRAS40 release. In the presence of Rheb and insulin, PRAS40 release is abolished by Akt inhibition without diminishing mTORC1 signaling. In conclusion, dissociation of PRAS40 from mTORC1 and enhanced mTORC1 substrate binding results from Akt and mTORC1 activation and makes little or no contribution to mTORC1 signaling, which rather is determined by Rheb activation of mTOR catalytic activity, through mechanisms that remain to be fully elucidated.
Figures
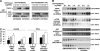
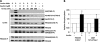



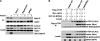

Similar articles
-
The proline-rich Akt substrate of 40 kDa (PRAS40) is a physiological substrate of mammalian target of rapamycin complex 1.J Biol Chem. 2007 Jul 13;282(28):20329-39. doi: 10.1074/jbc.M702636200. Epub 2007 May 21. J Biol Chem. 2007. PMID: 17517883 Free PMC article.
-
PRAS40 is a target for mammalian target of rapamycin complex 1 and is required for signaling downstream of this complex.J Biol Chem. 2007 Aug 24;282(34):24514-24. doi: 10.1074/jbc.M704406200. Epub 2007 Jun 29. J Biol Chem. 2007. PMID: 17604271
-
Hydrogen peroxide impairs insulin-stimulated assembly of mTORC1.Free Radic Biol Med. 2009 Jun 1;46(11):1500-9. doi: 10.1016/j.freeradbiomed.2009.03.001. Epub 2009 Mar 10. Free Radic Biol Med. 2009. PMID: 19281842 Free PMC article.
-
Proline-rich Akt substrate of 40kDa (PRAS40): a novel downstream target of PI3k/Akt signaling pathway.Cell Signal. 2012 Jan;24(1):17-24. doi: 10.1016/j.cellsig.2011.08.010. Epub 2011 Aug 31. Cell Signal. 2012. PMID: 21906675 Review.
-
Disentangling the signaling pathways of mTOR complexes, mTORC1 and mTORC2, as a therapeutic target in glioblastoma.Adv Biol Regul. 2022 Jan;83:100854. doi: 10.1016/j.jbior.2021.100854. Epub 2021 Dec 6. Adv Biol Regul. 2022. PMID: 34996736 Review.
Cited by
-
Alpha-ketoglutarate partially alleviates effects of high-fat high-fructose diet in mouse muscle.EXCLI J. 2023 Dec 5;22:1264-1277. doi: 10.17179/excli2023-6608. eCollection 2023. EXCLI J. 2023. PMID: 38234967 Free PMC article.
-
TBK1 Limits mTORC1 by Promoting Phosphorylation of Raptor Ser877.Sci Rep. 2019 Sep 17;9(1):13470. doi: 10.1038/s41598-019-49707-8. Sci Rep. 2019. PMID: 31530866 Free PMC article.
-
Phosphoproteomics reveals resveratrol-dependent inhibition of Akt/mTORC1/S6K1 signaling.J Proteome Res. 2014 Dec 5;13(12):5734-42. doi: 10.1021/pr500714a. Epub 2014 Oct 29. J Proteome Res. 2014. PMID: 25311616 Free PMC article.
-
Aescin Protects Neuron from Ischemia-Reperfusion Injury via Regulating the PRAS40/mTOR Signaling Pathway.Oxid Med Cell Longev. 2020 Sep 30;2020:7815325. doi: 10.1155/2020/7815325. eCollection 2020. Oxid Med Cell Longev. 2020. PMID: 33062146 Free PMC article.
-
The Role of the PI3K/Akt/mTOR Axis in Head and Neck Squamous Cell Carcinoma.Biomedicines. 2024 Jul 19;12(7):1610. doi: 10.3390/biomedicines12071610. Biomedicines. 2024. PMID: 39062182 Free PMC article. Review.
References
-
- Wullschleger S., Loewith R., Hall M. N. (2006) Cell 124, 471–484 - PubMed
-
- Hara K., Maruki Y., Long X., Yoshino K., Oshiro N., Hidayat S., Tokunaga C., Avruch J., Yonezawa K. (2002) Cell 110, 177–189 - PubMed
-
- Kim D. H., Sarbassov D. D., Ali S. M., King J. E., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2002) Cell 110, 163–175 - PubMed
-
- Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J. L., Bonenfant D., Oppliger W., Jenoe P., Hall M. N. (2002) Mol. Cell. 10, 457–468 - PubMed
-
- Wang L., Rhodes C. J., Lawrence J. C., Jr. (2006) J. Biol. Chem. 281, 24293–24303 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

