A BEN-domain-containing protein associates with heterochromatin and represses transcription
- PMID: 21914818
- PMCID: PMC4074281
- DOI: 10.1242/jcs.086603
A BEN-domain-containing protein associates with heterochromatin and represses transcription
Abstract
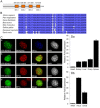
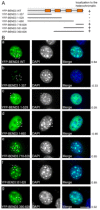
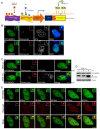
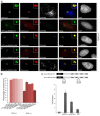
In eukaryotes, higher order chromatin structure governs crucial cellular processes including DNA replication, transcription and post-transcriptional gene regulation. Specific chromatin-interacting proteins play vital roles in the maintenance of chromatin structure. We have identified BEND3, a quadruple BEN domain-containing protein that is highly conserved amongst vertebrates. BEND3 colocalizes with HP1 and H3 trimethylated at K9 at heterochromatic regions in mammalian cells. Using an in vivo gene locus, we have been able to demonstrate that BEND3 associates with the locus only when it is heterochromatic and dissociates upon activation of transcription. Furthermore, tethering BEND3 inhibits transcription from the locus, indicating that BEND3 is involved in transcriptional repression through its interaction with histone deacetylases and Sall4, a transcription repressor. We further demonstrate that BEND3 is SUMOylated and that such modifications are essential for its role in transcriptional repression. Finally, overexpression of BEND3 causes premature chromatin condensation and extensive heterochromatinization, resulting in cell cycle arrest. Taken together, our data demonstrate the role of a novel heterochromatin-associated protein in transcriptional repression.
Figures


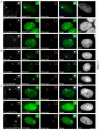
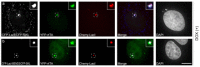
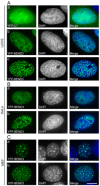
References
-
- Bannister A. J., Zegerman P., Partridge J. F., Miska E. A., Thomas J. O., Allshire R. C., Kouzarides T. (2001). Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410, 120-124 - PubMed
-
- Bayer P., Arndt A., Metzger S., Mahajan R., Melchior F., Jaenicke R., Becker J. (1998). Structure determination of the small ubiquitin-related modifier SUMO-1. J. Mol. Biol. 280, 275-286 - PubMed
-
- Berg J. M. (1990). Zinc finger domains: hypotheses and current knowledge. Annu. Rev. Biophys. Biophys. Chem. 19, 405-421 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

