Coupled pre-mRNA and mRNA dynamics unveil operational strategies underlying transcriptional responses to stimuli
- PMID: 21915116
- PMCID: PMC3202801
- DOI: 10.1038/msb.2011.62
Coupled pre-mRNA and mRNA dynamics unveil operational strategies underlying transcriptional responses to stimuli
Abstract
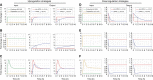
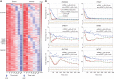
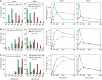
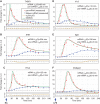
Transcriptional responses to extracellular stimuli involve tuning the rates of transcript production and degradation. Here, we show that the time-dependent profiles of these rates can be inferred from simultaneous measurements of precursor mRNA (pre-mRNA) and mature mRNA profiles. Transcriptome-wide measurements demonstrate that genes with similar mRNA profiles often exhibit marked differences in the amplitude and onset of their production rate. The latter is characterized by a large dynamic range, with a group of genes exhibiting an unexpectedly strong transient production overshoot, thereby accelerating their induction and, when combined with time-dependent degradation, shaping transient responses with precise timing and amplitude.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- Alon U (2007) An Introduction to Systems Biology—Designs Principles of Biological Circuits. Boca Raton, FL: Chapman and Hall/CRC
-
- Amit I, Citri A, Shay T, Lu Y, Katz M, Zhang F, Tarcic G, Siwak D, Lahad J, Jacob-Hirsch J, Amariglio N, Vaisman N, Segal E, Rechavi G, Alon U, Mills GB, Domany E, Yarden Y (2007) A module of negative feedback regulators defines growth factor signaling. Nat Genet 39: 503–512 - PubMed
-
- Amit I, Garber M, Chevrier N, Leite AP, Donner Y, Eisenhaure T, Guttman M, Grenier JK, Li W, Zuk O, Schubert LA, Birditt B, Shay T, Goren A, Zhang X, Smith Z, Deering R, McDonald RC, Cabili M, Bernstein BE et al. (2009) Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science 326: 257–263 - PMC - PubMed
-
- Avraham R, Sas-Chen A, Manor O, Steinfeld I, Shalgi R, Tarcic G, Bossel N, Zeisel A, Amit I, Zwang Y, Enerly E, Russnes HG, Biagioni F, Mottolese M, Strano S, Blandino G, Borresen-Dale AL, Pilpel Y, Yakhini Z, Segal E et al. (2010) EGF decreases the abundance of microRNAs that restrain oncogenic transcription factors. Sci Signal 3: ra43. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

