In situ AFM study of amelogenin assembly and disassembly dynamics on charged surfaces provides insights on matrix protein self-assembly
- PMID: 21916473
- PMCID: PMC3427831
- DOI: 10.1021/ja206849c
In situ AFM study of amelogenin assembly and disassembly dynamics on charged surfaces provides insights on matrix protein self-assembly
Abstract
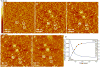
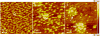
Because self-assembly of matrix proteins is a key step in hard tissue mineralization, developing an understanding of the assembly pathways and underlying mechanisms is likely to be important for successful hard tissue engineering. While many studies of matrix protein assembly have been performed on bulk solutions, in vivo these proteins are likely to be in contact with charged biological surfaces composed of lipids, proteins, or minerals. Here we report the results of an in situ atomic force microscopy (AFM) study of self-assembly by amelogenin--the principal protein of the extracellular matrix in developing enamel--in contact with two different charged substrates: hydrophilic negatively charged bare mica and positively charged 3-aminopropyl triethoxysilane (APS) silanized mica. First we demonstrate an AFM-based protocol for determining the size of both amelogenin monomers and oligomers. Using this protocol, we find that, although amelogenin exists primarily as ~26 nm in diameter nanospheres in bulk solution at a pH of 8.0 studied by dynamic light scattering, it behaves dramatically differently upon interacting with charged substrates at the same pH and exhibits complex substrate-dependent assembly pathways and dynamics. On positively charged APS-treated mica surfaces, amelogenin forms a relatively uniform population of decameric oligomers, which then transform into two main populations: higher-order assemblies of oligomers and amelogenin monomers, while on negatively charged bare mica surfaces, it forms a film of monomers that exhibits tip-induced desorption and patterning. The present study represents a successful attempt to identify the size of amelogenin oligomers and to directly monitor assembly and disassembly dynamics on surfaces. The findings have implications for amelogenin-controlled calcium phosphate mineralization in vitro and may offer new insights into in vivo self-assembly of matrix proteins as well as their control over hard tissue formation.
Figures

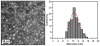



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

