Viruses and sialic acids: rules of engagement
- PMID: 21917445
- PMCID: PMC3189341
- DOI: 10.1016/j.sbi.2011.08.009
Viruses and sialic acids: rules of engagement
Abstract
Viral infections are initiated by specific attachment of a virus particle to receptors at the surface of the host cell. For many viruses, these receptors are glycans that are linked to either a protein or a lipid. Glycans terminating in sialic acid and its derivatives serve as receptors for a large number of viruses, including several human pathogens. In combination with glycan array analyses, structural analyses of complexes of viruses with sialylated oligosaccharides have provided insights into the parameters that underlie each interaction. Here, we compare the currently available structural data on viral attachment proteins in complex with sialic acid and its variants. The objective is to define common parameters of recognition and to provide a platform for understanding the determinants of specificity. This information could be of use for the prediction of the location of sialic acid binding sites in viruses for which structural information is still lacking. An improved understanding of the principles that govern the recognition of sialic acid and sialylated oligosaccharides would also advance efforts to develop efficient antiviral agents.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures
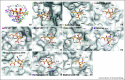
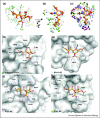
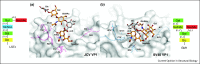

References
-
- Villar E., Barroso I.M. Role of sialic acid-containing molecules in paramyxovirus entry into the host cell: a minireview. Glycoconj J. 2006;23:5–17. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

