Novel pentameric structure of the diarrhea-inducing region of the rotavirus enterotoxigenic protein NSP4
- PMID: 21917949
- PMCID: PMC3209389
- DOI: 10.1128/JVI.00349-11
Novel pentameric structure of the diarrhea-inducing region of the rotavirus enterotoxigenic protein NSP4
Abstract
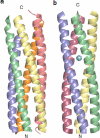
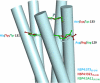
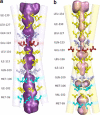
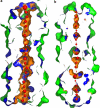
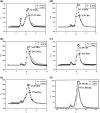
A novel pentameric structure which differs from the previously reported tetrameric form of the diarrhea-inducing region of the rotavirus enterotoxin NSP4 is reported here. A significant feature of this pentameric form is the absence of the calcium ion located in the core region of the tetrameric structures. The lysis of cells, the crystallization of the region spanning residues 95 to 146 of NSP4 (NSP4(95-146)) of strain ST3 (ST3:NSP4(95-146)) at acidic pH, and comparative studies of the recombinant purified peptide under different conditions by size-exclusion chromatography (SEC) and of the crystal structures suggested pH-, Ca(2+)-, and protein concentration-dependent oligomeric transitions in the peptide. Since the NSP4(95-146) mutant lacks the N-terminal amphipathic domain (AD) and most of the C-terminal flexible region (FR), to demonstrate that the pentameric transition is not a consequence of the lack of the N- and C-terminal regions, glutaraldehyde cross-linking of the ΔN72 and ΔN94 mutant proteins, which contain or lack the AD, respectively, but possess the complete C-terminal FR, was carried out. The results indicate the presence of pentamers in preparations of these longer mutants. Detailed SEC analyses of ΔN94 prepared under different conditions, however, revealed protein concentration-dependent but metal ion- and pH-independent pentamer accumulation at high concentrations which dissociated into tetramers and lower oligomers at low protein concentrations. While calcium appeared to stabilize the tetramer, magnesium in particular stabilized the dimer. ΔN72 existed primarily in the multimeric form under all conditions. These findings of a calcium-free NSP4 pentamer and its concentration-dependent and largely calcium-independent oligomeric transitions open up a new dimension in an understanding of the structural basis of its multitude of functions.
Figures
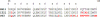
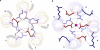

Similar articles
-
Structural plasticity of the coiled-coil domain of rotavirus NSP4.J Virol. 2014 Dec;88(23):13602-12. doi: 10.1128/JVI.02227-14. Epub 2014 Sep 17. J Virol. 2014. PMID: 25231315 Free PMC article.
-
Structure of the extended diarrhea-inducing domain of rotavirus enterotoxigenic protein NSP4.Arch Virol. 2007;152(5):847-59. doi: 10.1007/s00705-006-0921-x. Epub 2007 Jan 31. Arch Virol. 2007. PMID: 17265103
-
New tetrameric forms of the rotavirus NSP4 with antiparallel helices.Arch Virol. 2018 Jun;163(6):1531-1547. doi: 10.1007/s00705-018-3753-6. Epub 2018 Feb 17. Arch Virol. 2018. PMID: 29455326
-
How do the rotavirus NSP4 and bacterial enterotoxins lead differently to diarrhea?Virol J. 2007 Mar 21;4:31. doi: 10.1186/1743-422X-4-31. Virol J. 2007. PMID: 17376232 Free PMC article. Review.
-
Microbes and microbial toxins: paradigms for microbial-mucosal interactions. VIII. Pathological consequences of rotavirus infection and its enterotoxin.Am J Physiol Gastrointest Liver Physiol. 2001 Aug;281(2):G303-10. doi: 10.1152/ajpgi.2001.281.2.G303. Am J Physiol Gastrointest Liver Physiol. 2001. PMID: 11447008 Review.
Cited by
-
Genetic divergence of rotavirus nonstructural protein 4 results in distinct serogroup-specific viroporin activity and intracellular punctate structure morphologies.J Virol. 2012 May;86(9):4921-34. doi: 10.1128/JVI.06759-11. Epub 2012 Feb 22. J Virol. 2012. PMID: 22357281 Free PMC article.
-
The Rotavirus NSP4 Viroporin Domain is a Calcium-conducting Ion Channel.Sci Rep. 2017 Mar 3;7:43487. doi: 10.1038/srep43487. Sci Rep. 2017. PMID: 28256607 Free PMC article.
-
Rotavirus non-structural proteins: structure and function.Curr Opin Virol. 2012 Aug;2(4):380-8. doi: 10.1016/j.coviro.2012.06.003. Epub 2012 Jul 11. Curr Opin Virol. 2012. PMID: 22789743 Free PMC article. Review.
-
Efficient intracellular delivery of proteins by a multifunctional chimaeric peptide in vitro and in vivo.Nat Commun. 2021 Aug 26;12(1):5131. doi: 10.1038/s41467-021-25448-z. Nat Commun. 2021. PMID: 34446736 Free PMC article.
-
Modeling of the Ebola virus delta peptide reveals a potential lytic sequence motif.Viruses. 2015 Jan 20;7(1):285-305. doi: 10.3390/v7010285. Viruses. 2015. PMID: 25609303 Free PMC article.
References
-
- Adams P. D., et al. 2002. Crystallographic structure determination. Acta Crystallogr. D 58:1948–1954 - PubMed
-
- Ball J. M., Tian P., Zeng C. Q.-Y., Morris A. P., Estes M. K. 1996. Age-dependent diarrhea induced by a rotaviral nonstructural glycoprotein. Science 272:101–104 - PubMed
-
- Bas D. C., Rogers D. M., Jensen J. H. 2008. Very fast prediction and rationalization of pKa values for protein-ligand complexes. Proteins 73:765–783 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

