Differential reactivity of germ line allelic variants of a broadly neutralizing HIV-1 antibody to a gp41 fusion intermediate conformation
- PMID: 21917975
- PMCID: PMC3209283
- DOI: 10.1128/JVI.05680-11
Differential reactivity of germ line allelic variants of a broadly neutralizing HIV-1 antibody to a gp41 fusion intermediate conformation
Abstract
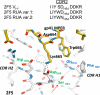
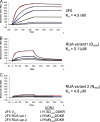
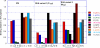
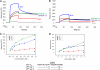
Genetic factors, as well as antigenic stimuli, can influence antibody repertoire formation. Moreover, the affinity of antigen for unmutated naïve B cell receptors determines the threshold for activation of germinal center antibody responses. The gp41 2F5 broadly neutralizing antibody (bNAb) uses the V(H)2-5 gene, which has 10 distinct alleles that use either a heavy-chain complementarity-determining region 2 (HCDR2) aspartic acid (D(H54)) or an HCDR2 asparagine (N(H54)) residue. The 2F5 HCDR2 D(H54) residue has been shown to form a salt bridge with gp41 (665)K; the V(H)2-5 germ line allele variant containing N(H54) cannot do so and thus should bind less avidly to gp41. Thus, the induction of 2F5 bNAb is dependent on both genetic and structural factors that could affect antigen affinity of unmutated naïve B cell receptors. Here, we studied allelic variants of the V(H)2-5 inferred germ line forms of the HIV-1 gp41 bNAb 2F5 for their antigen binding affinities to gp41 linear peptide and conformational protein antigens. Both V(H)2-5 2F5 inferred germ line variants bound to gp41 peptides and protein, including the fusion intermediate protein mimic, although more weakly than the mature 2F5 antibody. As predicted, the affinity of the N(H54) variant for fusion-intermediate conformation was an order of magnitude lower than that of the D(H54) V(H)2-5 germ line antibody, demonstrating that allelic variants of 2F5 germ line antibodies differentially bind to gp41. Thus, these data demonstrate a genetically determined trait that may affect host responses to HIV-1 envelope epitopes recognized by broadly neutralizing antibodies and has implications for unmutated ancestor-based immunogen design.
Figures

References
-
- Berman J. E., Alt F. W. 1990. Human heavy chain variable region gene diversity, organization, and expression. Int. Rev. Immunol. 5: 203–214 - PubMed
-
- Cardoso R. M., et al. 2005. Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity 22: 163–173 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

