Mutations in the membrane-proximal region of the influenza A virus M2 protein cytoplasmic tail have modest effects on virus replication
- PMID: 21917980
- PMCID: PMC3209349
- DOI: 10.1128/JVI.05970-11
Mutations in the membrane-proximal region of the influenza A virus M2 protein cytoplasmic tail have modest effects on virus replication
Abstract
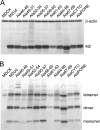
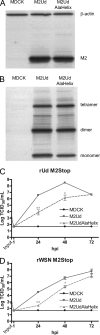
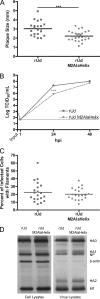
Influenza A virus encodes M2, a proton channel that has been shown to be important during virus entry and assembly. In order to systematically investigate the role of the membrane-proximal residues in the M2 cytoplasmic tail in virus replication, we utilized scanning and directed alanine mutagenesis in combination with transcomplementation assays and recombinant viruses. The membrane-proximal residues 46 to 69 tolerated numerous mutations, with little, if any, effect on virus replication, suggesting that protein structure rather than individual amino acid identity in this region may be critical for M2 protein function.
Figures



References
-
- Bourmakina S. V., Garcia-Sastre A. 2003. Reverse genetics studies on the filamentous morphology of influenza A virus. J. Gen. Virol. 84:517–527 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

