The nucleosome remodeling factor
- PMID: 21920360
- PMCID: PMC4839296
- DOI: 10.1016/j.febslet.2011.09.003
The nucleosome remodeling factor
Abstract
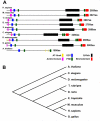
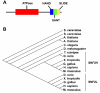
An essential component of the chromatin remodeling machinery is NURF (Nucleosome Remodeling Factor), the founding member of the ISWI family of chromatin remodeling complexes. In vertebrates and invertebrates alike, NURF has many important functions in chromatin biology including regulating transcription, establishing boundary elements, and promoting higher order chromatin structure. Since NURF is essential to many aspects of chromatin biology, knowledge of its function is required to fully understand how the genome is regulated. This review will summarize what is currently known of its biological functions, conservation in the most prominent model organisms, biochemical functions as a nucleosome remodeling enzyme, and its possible relevance to human cancer.
Copyright © 2011 Federation of European Biochemical Societies. Published by Elsevier B.V. All rights reserved.
Figures



References
-
- Luger K, et al. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–260. - PubMed
-
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009;78:273–304. - PubMed
-
- Tsukiyama T, Becker PB, Wu C. ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature. 1994;367(6463):525–532. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

