Elucidation of the biosynthetic pathway for Okenone in Thiodictyon sp. CAD16 leads to the discovery of two novel carotene ketolases
- PMID: 21921032
- PMCID: PMC3207450
- DOI: 10.1074/jbc.M111.280131
Elucidation of the biosynthetic pathway for Okenone in Thiodictyon sp. CAD16 leads to the discovery of two novel carotene ketolases
Abstract
Okenone is a unique ketocarotenoid found in many purple sulfur bacteria; it is important because of its unique light absorption and photoprotection properties. Okenane, a compound formed by diagenetic reduction of okenone, is an important biomarker in geochemical analyses of sedimentary rocks. Despite its ecological and biogeochemical importance, the biochemical pathway for okenone synthesis has not yet been fully described. The genome sequence of an okenone-producing organism, Thiodictyon sp. strain CAD16, revealed four genes whose predicted proteins had strong sequence similarity to enzymes known to produce ψ-end group modifications of carotenoids in proteobacteria. These four genes encoded homologs of a 1,2-carotenoid hydratase (CrtC), an O-methyltransferase (CrtF), and two paralogs of carotenoid 3,4-desaturases (CrtD). Expression studies in lycopene- or neurosporene-producing strains of Escherichia coli confirmed the functions of crtC and crtF, but the crtD paralogs encoded enzymes with previously undescribed functions. One enzyme, CruS, was only distantly related to CrtD desaturases, was bifunctional, and performed a 3,4-desaturation and introduced a C-2 keto group into neurosporene derivatives in the presence of dioxygen. The enzyme encoded by the other crtD paralog also represents a new enzyme in carotenogenesis and was named cruO. CruO encodes the C-4/4' ketolase uniquely required for okenone biosynthesis. The identification of CruO and the demonstration of its biochemical activity complete the elucidation of the biosynthetic pathway for okenone and other related ketocarotenoids.
Figures


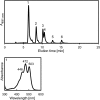
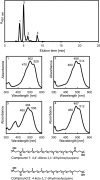

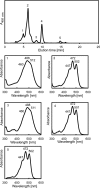
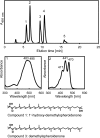


References
-
- Imhoff J. (2006) in The Prokaryotes (Dworkin M., Falkow S., Rosenberg E., Schleifer K.-H., Stackebrandt E. eds) pp. 874–886, Springer, New York
-
- Imhoff J. (2006) in The Prokaryotes (Dworkin M., Falkow S., Rosenberg E., Schleifer K.-H., Stackebrandt E. eds) pp. 41–64, Springer, New York
-
- Imhoff J. (2006) in The Prokaryotes (Dworkin M., Falkow S., Rosenberg E., Schleifer K.-H., Stackebrandt E. eds) pp. 593–601, Springer, New York
-
- Imhoff J. F. (2006) in The Prokaryotes (Dworkin M., Falkow S., Rosenberg E., Schleifer K.-H., Stackebrandt E. eds) pp. 846–873, Springer, New York
-
- Madigan M. T., Jung D. O. (2009) in The Purple Phototrophic Bacteria (Hunter C. N., Daldal F., Thurnauer M. C., Beatty J. T. eds) pp. 1–15, Springer, Dordrecht, The Netherlands
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

