Experimental design and statistical rigor in phylogenomics of horizontal and endosymbiotic gene transfer
- PMID: 21923904
- PMCID: PMC3190393
- DOI: 10.1186/1471-2148-11-259
Experimental design and statistical rigor in phylogenomics of horizontal and endosymbiotic gene transfer
Abstract
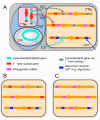
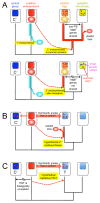
A growing number of phylogenomic investigations from diverse eukaryotes are examining conflicts among gene trees as evidence of horizontal gene transfer. If multiple foreign genes from the same eukaryotic lineage are found in a given genome, it is increasingly interpreted as concerted gene transfers during a cryptic endosymbiosis in the organism's evolutionary past, also known as "endosymbiotic gene transfer" or EGT. A number of provocative hypotheses of lost or serially replaced endosymbionts have been advanced; to date, however, these inferences largely have been post-hoc interpretations of genomic-wide conflicts among gene trees. With data sets as large and complex as eukaryotic genome sequences, it is critical to examine alternative explanations for intra-genome phylogenetic conflicts, particularly how much conflicting signal is expected from directional biases and statistical noise. The availability of genome-level data both permits and necessitates phylogenomics that test explicit, a priori predictions of horizontal gene transfer, using rigorous statistical methods and clearly defined experimental controls.
Figures
Similar articles
-
Endosymbiotic gene transfer from prokaryotic pangenomes: Inherited chimerism in eukaryotes.Proc Natl Acad Sci U S A. 2015 Aug 18;112(33):10139-46. doi: 10.1073/pnas.1421385112. Epub 2015 Mar 2. Proc Natl Acad Sci U S A. 2015. PMID: 25733873 Free PMC article.
-
Phylogenomic test of the hypotheses for the evolutionary origin of eukaryotes.Mol Biol Evol. 2014 Apr;31(4):832-45. doi: 10.1093/molbev/mst272. Epub 2014 Jan 7. Mol Biol Evol. 2014. PMID: 24398320 Free PMC article.
-
Recent events dominate interdomain lateral gene transfers between prokaryotes and eukaryotes and, with the exception of endosymbiotic gene transfers, few ancient transfer events persist.Philos Trans R Soc Lond B Biol Sci. 2015 Sep 26;370(1678):20140324. doi: 10.1098/rstb.2014.0324. Philos Trans R Soc Lond B Biol Sci. 2015. PMID: 26323756 Free PMC article.
-
Witnessing Genome Evolution: Experimental Reconstruction of Endosymbiotic and Horizontal Gene Transfer.Annu Rev Genet. 2017 Nov 27;51:1-22. doi: 10.1146/annurev-genet-120215-035329. Epub 2017 Aug 28. Annu Rev Genet. 2017. PMID: 28846455 Review.
-
The phylogenomics of protein structures: The backstory.Biochimie. 2015 Dec;119:284-302. doi: 10.1016/j.biochi.2015.07.027. Epub 2015 Jul 31. Biochimie. 2015. PMID: 26234735 Review.
Cited by
-
Controversies in modern evolutionary biology: the imperative for error detection and quality control.BMC Genomics. 2012 Jan 4;13:5. doi: 10.1186/1471-2164-13-5. BMC Genomics. 2012. PMID: 22217008 Free PMC article.
-
Reevaluating the green contribution to diatom genomes.Genome Biol Evol. 2012;4(7):683-8. doi: 10.1093/gbe/evs053. Epub 2012 Jun 7. Genome Biol Evol. 2012. PMID: 22684208 Free PMC article.
-
The eukaryotic tree of life from a global phylogenomic perspective.Cold Spring Harb Perspect Biol. 2014 May 1;6(5):a016147. doi: 10.1101/cshperspect.a016147. Cold Spring Harb Perspect Biol. 2014. PMID: 24789819 Free PMC article. Review.
-
Genomes of Stigonematalean cyanobacteria (subsection V) and the evolution of oxygenic photosynthesis from prokaryotes to plastids.Genome Biol Evol. 2013;5(1):31-44. doi: 10.1093/gbe/evs117. Genome Biol Evol. 2013. PMID: 23221676 Free PMC article.
-
Nitrile hydratase genes are present in multiple eukaryotic supergroups.PLoS One. 2012;7(4):e32867. doi: 10.1371/journal.pone.0032867. Epub 2012 Apr 10. PLoS One. 2012. PMID: 22505998 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

