A new class of IMP dehydrogenase with a role in self-resistance of mycophenolic acid producing fungi
- PMID: 21923907
- PMCID: PMC3184278
- DOI: 10.1186/1471-2180-11-202
A new class of IMP dehydrogenase with a role in self-resistance of mycophenolic acid producing fungi
Abstract
Background: Many secondary metabolites produced by filamentous fungi have potent biological activities, to which the producer organism must be resistant. An example of pharmaceutical interest is mycophenolic acid (MPA), an immunosuppressant molecule produced by several Penicillium species. The target of MPA is inosine-5'-monophosphate dehydrogenase (IMPDH), which catalyses the rate limiting step in the synthesis of guanine nucleotides. The recent discovery of the MPA biosynthetic gene cluster from Penicillium brevicompactum revealed an extra copy of the IMPDH-encoding gene (mpaF) embedded within the cluster. This finding suggests that the key component of MPA self resistance is likely based on the IMPDH encoded by mpaF.
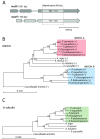
Results: In accordance with our hypothesis, heterologous expression of mpaF dramatically increased MPA resistance in a model fungus, Aspergillus nidulans, which does not produce MPA. The growth of an A. nidulans strain expressing mpaF was only marginally affected by MPA at concentrations as high as 200 μg/ml. To further substantiate the role of mpaF in MPA resistance, we searched for mpaF orthologs in six MPA producer/non-producer strains from Penicillium subgenus Penicillium. All six strains were found to hold two copies of IMPDH. A cladistic analysis based on the corresponding cDNA sequences revealed a novel group constituting mpaF homologs. Interestingly, a conserved tyrosine residue in the original class of IMPDHs is replaced by a phenylalanine residue in the new IMPDH class.
Conclusions: We identified a novel variant of the IMPDH-encoding gene in six different strains from Penicillium subgenus Penicillium. The novel IMPDH variant from MPA producer P. brevicompactum was shown to confer a high degree of MPA resistance when expressed in a non-producer fungus. Our study provides a basis for understanding the molecular mechanism of MPA resistance and has relevance for biotechnological and pharmaceutical applications.
Figures


References
-
- Weber G, Nakamura H, Natsumeda Y, Szekeres T, Nagai M. Regulation of GTP biosynthesis. Advances in enzyme regulation. 1992;32:57–69. - PubMed
-
- Frisvad JC, Smedsgaard JR, Larsen TO, Samson RA. Mycotoxins, drugs and other extrolites produced by species in Penicillium subgenus Penicillium. Stud Mycol. 2004;49:201–41.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

