Molecular genetic analysis of circadian timekeeping in Drosophila
- PMID: 21924977
- PMCID: PMC4108082
- DOI: 10.1016/B978-0-12-387690-4.00005-2
Molecular genetic analysis of circadian timekeeping in Drosophila
Abstract
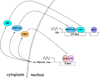
A genetic screen for mutants that alter circadian rhythms in Drosophila identified the first clock gene-the period (per) gene. The per gene is a central player within a transcriptional feedback loop that represents the core mechanism for keeping circadian time in Drosophila and other animals. The per feedback loop, or core loop, is interlocked with the Clock (Clk) feedback loop, but whether the Clk feedback loop contributes to circadian timekeeping is not known. A series of distinct molecular events are thought to control transcriptional feedback in the core loop. The time it takes to complete these events should take much less than 24h, thus delays must be imposed at different steps within the core loop. As new clock genes are identified, the molecular mechanisms responsible for these delays have been revealed in ever-increasing detail and provide an in-depth accounting of how transcriptional feedback loops keep circadian time. The phase of these feedback loops shifts to maintain synchrony with environmental cycles, the most reliable of which is light. Although a great deal is known about cell-autonomous mechanisms of light-induced phase shifting by CRYPTOCHROME (CRY), much less is known about non-cell autonomous mechanisms. CRY mediates phase shifts through an uncharacterized mechanism in certain brain oscillator neurons and carries out a dual role as a photoreceptor and transcription factor in other tissues. Here, I review how transcriptional feedback loops function to keep time in Drosophila, how they impose delays to maintain a 24-h cycle, and how they maintain synchrony with environmental light:dark cycles. The transcriptional feedback loops that keep time in Drosophila are well conserved in other animals, thus what we learn about these loops in Drosophila should continue to provide insight into the operation of analogous transcriptional feedback loops in other animals.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures


References
-
- Ahmad M, Cashmore AR. HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature. 1993;366:162–166. - PubMed
-
- Akten B, Jauch E, Genova GK, Kim EY, Edery I, Raabe T, Jackson FR. A role for CK2 in the Drosophila circadian oscillator. Nat Neurosci. 2003;6:251–257. - PubMed
-
- Allada R, White NE, So WV, Hall JC, Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

