Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification
- PMID: 21925322
- PMCID: PMC3176443
- DOI: 10.1016/j.cell.2011.08.008
Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification
Abstract
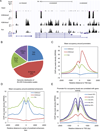
We report the identification of 67 previously undescribed histone modifications, increasing the current number of known histone marks by about 70%. We further investigated one of the marks, lysine crotonylation (Kcr), confirming that it represents an evolutionarily-conserved histone posttranslational modification. The unique structure and genomic localization of histone Kcr suggest that it is mechanistically and functionally different from histone lysine acetylation (Kac). Specifically, in both human somatic and mouse male germ cell genomes, histone Kcr marks either active promoters or potential enhancers. In male germinal cells immediately following meiosis, Kcr is enriched on sex chromosomes and specifically marks testis-specific genes, including a significant proportion of X-linked genes that escape sex chromosome inactivation in haploid cells. These results therefore dramatically extend the repertoire of histone PTM sites and designate Kcr as a specific mark of active sex chromosome-linked genes in postmeiotic male germ cells.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures





Comment in
-
Chromatin: a haul of new histone modifications.Nat Rev Genet. 2011 Sep 27;12(11):744. doi: 10.1038/nrg3086. Nat Rev Genet. 2011. PMID: 21946920 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

