Prospects for riboswitch discovery and analysis
- PMID: 21925376
- PMCID: PMC4140403
- DOI: 10.1016/j.molcel.2011.08.024
Prospects for riboswitch discovery and analysis
Abstract
An expanding number of metabolite-binding riboswitch classes are being discovered in the noncoding portions of bacterial genomes. Findings over the last decade indicate that bacteria commonly use these RNA genetic elements as regulators of metabolic pathways and as mediators of changes in cell physiology. Some riboswitches are surprisingly complex, and they rival protein factors in their structural and functional sophistication. Each new riboswitch discovery expands our knowledge of the biochemical capabilities of RNA, and some give rise to new questions that require additional study to be addressed. Some of the greatest prospects for riboswitch research and some of the more interesting mysteries are discussed in this review.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
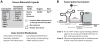


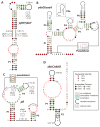

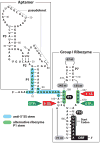
References
-
- Ames TD, Breaker RR. Bacterial riboswitch discovery and analysis. In: Meyer G, editor. The Chemical Biology of Nucleic Acids. John Wiley and Sons, Ltd; 2010. pp. 433–452.
-
- Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

