Regulation by small RNAs in bacteria: expanding frontiers
- PMID: 21925377
- PMCID: PMC3176440
- DOI: 10.1016/j.molcel.2011.08.022
Regulation by small RNAs in bacteria: expanding frontiers
Abstract
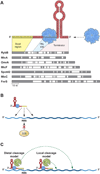
Research on the discovery and characterization of small, regulatory RNAs in bacteria has exploded in recent years. These sRNAs act by base pairing with target mRNAs with which they share limited or extended complementarity, or by modulating protein activity, in some cases by mimicking other nucleic acids. Mechanistic insights into how sRNAs bind mRNAs and proteins, how they compete with each other, and how they interface with ribonucleases are active areas of discovery. Current work also has begun to illuminate how sRNAs modulate expression of distinct regulons and key transcription factors, thus integrating sRNA activity into extensive regulatory networks. In addition, the application of RNA deep sequencing has led to reports of hundreds of additional sRNA candidates in a wide swath of bacterial species. Most importantly, recent studies have served to clarify the abundance of remaining questions about how, when, and why sRNA-mediated regulation is of such importance to bacterial lifestyles.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures

References
-
- Babitzke P, Romeo T. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr. Opin. Microbiol. 2007;10:156–163. - PubMed
-
- Backofen R, Hess WR. Computational prediction of sRNAs and their targets in bacteria. RNA Biol. 2010;7:33–42. - PubMed
-
- Balbontín R, Fiorini F, Figueroa-Bossi N, Casadesús J, Bossi L. Recognition of heptameric seed sequence underlies multi-target regulation by RybB small RNA in Salmonella enterica. Mol. Microbiol. 2010;78:380–394. - PubMed
-
- Barrick JE, Yu DS, Yoon SH, Jeong H, Oh TK, Schneider D, Lenski RE, Kim JF. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 2009;461:1243–1247. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

