Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants
- PMID: 21925378
- PMCID: PMC3176435
- DOI: 10.1016/j.molcel.2011.07.024
Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants
Abstract
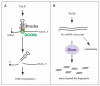
Since the establishment of a canonical animal microRNA biogenesis pathway driven by the RNase III enzymes Drosha and Dicer, an unexpected variety of alternative mechanisms that generate functional microRNAs have emerged. We review here the many Drosha-independent and Dicer-independent microRNA biogenesis strategies characterized over the past few years. Beyond reflecting the flexibility of small RNA machineries, the existence of noncanonical pathways has consequences for interpreting mutants in the core microRNA machinery. Such mutants are commonly used to assess the consequences of "total" microRNA loss, and indeed, they exhibit many overall phenotypic similarities. Nevertheless, ongoing studies reveal a growing number of settings in which alternative microRNA pathways contribute to distinct phenotypes among core microRNA biogenesis mutants.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

