CodY-mediated regulation of guanosine uptake in Bacillus subtilis
- PMID: 21926227
- PMCID: PMC3209203
- DOI: 10.1128/JB.05899-11
CodY-mediated regulation of guanosine uptake in Bacillus subtilis
Abstract
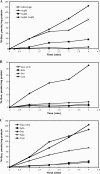
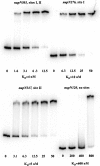
CodY is a global transcriptional regulator known to control expression of more than 100 genes and operons in Bacillus subtilis. Some of the most strongly repressed targets of CodY, the nupNOPQ (formerly, yufNOPQ) genes, were found to encode a guanosine transporter. Using DNase I footprinting experiments, we identified two high-affinity CodY-binding sites in the regulatory region of the nupN gene. The two sites are located 50 bp upstream and 163 bp downstream of the transcription start site. The downstream site was responsible for 6- to 8-fold nupN repression in the absence of the upstream site. When the upstream site was intact, however, only a minor contribution of the downstream site to nupN regulation could be detected under the conditions tested. Both sites contained 15-bp CodY-binding motifs with two mismatches each with respect to the consensus sequence, AATTTTCWGTTTTAA. However, the experimentally determined binding sites included additional sequences flanking the 15-bp CodY-binding motifs. An additional version of the 15-bp CodY-binding motif, with 5 mismatches with respect to the consensus but essential for efficient regulation by CodY, was found within the upstream site. The presence of multiple 15-bp motifs may be a common feature of CodY-binding sites.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

