RF1 knockout allows ribosomal incorporation of unnatural amino acids at multiple sites
- PMID: 21926996
- PMCID: PMC3201715
- DOI: 10.1038/nchembio.657
RF1 knockout allows ribosomal incorporation of unnatural amino acids at multiple sites
Abstract
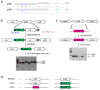
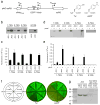
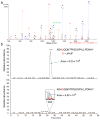
Stop codons have been exploited for genetic incorporation of unnatural amino acids (Uaas) in live cells, but their low incorporation efficiency, which is possibly due to competition from release factors, limits the power and scope of this technology. Here we show that the reportedly essential release factor 1 (RF1) can be knocked out from Escherichia coli by 'fixing' release factor 2 (RF2). The resultant strain JX33 is stable and independent, and it allows UAG to be reassigned from a stop signal to an amino acid when a UAG-decoding tRNA-synthetase pair is introduced. Uaas were efficiently incorporated at multiple UAG sites in the same gene without translational termination in JX33. We also found that amino acid incorporation at endogenous UAG codons is dependent on RF1 and mRNA context, which explains why E. coli tolerates apparent global suppression of UAG. JX33 affords a unique autonomous host for synthesizing and evolving new protein functions by enabling Uaa incorporation at multiple sites.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

