Evaluation of residue-residue contact predictions in CASP9
- PMID: 21928322
- PMCID: PMC3226919
- DOI: 10.1002/prot.23160
Evaluation of residue-residue contact predictions in CASP9
Abstract
This work presents the results of the assessment of the intramolecular residue-residue contact predictions submitted to CASP9. The methodology for the assessment does not differ from that used in previous CASPs, with two basic evaluation measures being the precision in recognizing contacts and the difference between the distribution of distances in the subset of predicted contact pairs versus all pairs of residues in the structure. The emphasis is placed on the prediction of long-range contacts (i.e., contacts between residues separated by at least 24 residues along sequence) in target proteins that cannot be easily modeled by homology. Although there is considerable activity in the field, the current analysis reports no discernable progress since CASP8.
Copyright © 2011 Wiley-Liss, Inc.
Figures
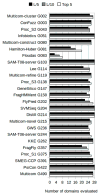
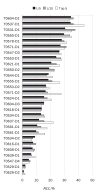
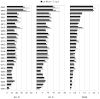
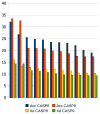
Similar articles
-
Evaluation of residue-residue contact prediction in CASP10.Proteins. 2014 Feb;82 Suppl 2(0 2):138-53. doi: 10.1002/prot.24340. Epub 2013 Aug 31. Proteins. 2014. PMID: 23760879 Free PMC article.
-
Evaluation of disorder predictions in CASP9.Proteins. 2011;79 Suppl 10(S10):107-18. doi: 10.1002/prot.23161. Epub 2011 Sep 16. Proteins. 2011. PMID: 21928402 Free PMC article.
-
Assessment of domain boundary predictions and the prediction of intramolecular contacts in CASP8.Proteins. 2009;77 Suppl 9:196-209. doi: 10.1002/prot.22554. Proteins. 2009. PMID: 19714769
-
Assessing the accuracy of contact and distance predictions in CASP14.Proteins. 2021 Dec;89(12):1888-1900. doi: 10.1002/prot.26248. Epub 2021 Oct 3. Proteins. 2021. PMID: 34595772 Free PMC article.
-
Improving accuracy of protein contact prediction using balanced network deconvolution.Proteins. 2015 Mar;83(3):485-96. doi: 10.1002/prot.24744. Epub 2015 Jan 24. Proteins. 2015. PMID: 25524593 Free PMC article.
Cited by
-
The MULTICOM toolbox for protein structure prediction.BMC Bioinformatics. 2012 Apr 30;13:65. doi: 10.1186/1471-2105-13-65. BMC Bioinformatics. 2012. PMID: 22545707 Free PMC article.
-
Sequence-based Gaussian network model for protein dynamics.Bioinformatics. 2014 Feb 15;30(4):497-505. doi: 10.1093/bioinformatics/btt716. Epub 2013 Dec 12. Bioinformatics. 2014. PMID: 24336646 Free PMC article.
-
NeBcon: protein contact map prediction using neural network training coupled with naïve Bayes classifiers.Bioinformatics. 2017 Aug 1;33(15):2296-2306. doi: 10.1093/bioinformatics/btx164. Bioinformatics. 2017. PMID: 28369334 Free PMC article.
-
Predicting three-dimensional structures of transmembrane domains of β-barrel membrane proteins.J Am Chem Soc. 2012 Jan 25;134(3):1775-81. doi: 10.1021/ja209895m. Epub 2012 Jan 12. J Am Chem Soc. 2012. PMID: 22148174 Free PMC article.
-
CONFOLD: Residue-residue contact-guided ab initio protein folding.Proteins. 2015 Aug;83(8):1436-49. doi: 10.1002/prot.24829. Epub 2015 Jun 6. Proteins. 2015. PMID: 25974172 Free PMC article.
References
-
- Niggemann M, Steipe B. Exploring local and non-local interactions for protein stability by structural motif engineering. J Mol Biol. 2000;296(1):181–195. - PubMed
-
- Gromiha MM, Selvaraj S. Inter-residue interactions in protein folding and stability. Prog Biophys Mol Biol. 2004;86(2):235–277. - PubMed
-
- Vendruscolo M, Kussell E, Domany E. Recovery of protein structure from contact maps. Fold Des. 1997;2(5):295–306. - PubMed
-
- Bohr J, Bohr H, Brunak S, Cotterill RM, Fredholm H, Lautrup B, Petersen SB. Protein structures from distance inequalities. J Mol Biol. 1993;231(3):861–869. - PubMed
-
- Pollastri G, Vullo A, Frasconi P, Baldi P. Modular DAG-RNN architectures for assembling coarse protein structures. J Comput Biol. 2006;13(3):631–650. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

