Families of transposable elements, population structure and the origin of species
- PMID: 21929767
- PMCID: PMC3183009
- DOI: 10.1186/1745-6150-6-44
Families of transposable elements, population structure and the origin of species
Abstract
Background: Eukaryotic genomes harbor diverse families of repetitive DNA derived from transposable elements (TEs) that are able to replicate and insert into genomic DNA. The biological role of TEs remains unclear, although they have profound mutagenic impact on eukaryotic genomes and the origin of repetitive families often correlates with speciation events. We present a new hypothesis to explain the observed correlations based on classical concepts of population genetics.
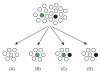
Presentation of the hypothesis: The main thesis presented in this paper is that the TE-derived repetitive families originate primarily by genetic drift in small populations derived mostly by subdivisions of large populations into subpopulations. We outline the potential impact of the emerging repetitive families on genetic diversification of different subpopulations, and discuss implications of such diversification for the origin of new species.
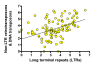
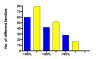
Testing the hypothesis: Several testable predictions of the hypothesis are examined. First, we focus on the prediction that the number of diverse families of TEs fixed in a representative genome of a particular species positively correlates with the cumulative number of subpopulations (demes) in the historical metapopulation from which the species has emerged. Furthermore, we present evidence indicating that human AluYa5 and AluYb8 families might have originated in separate proto-human subpopulations. We also revisit prior evidence linking the origin of repetitive families to mammalian phylogeny and present additional evidence linking repetitive families to speciation based on mammalian taxonomy. Finally, we discuss evidence that mammalian orders represented by the largest numbers of species may be subject to relatively recent population subdivisions and speciation events.
Implications of the hypothesis: The hypothesis implies that subdivision of a population into small subpopulations is the major step in the origin of new families of TEs as well as of new species. The origin of new subpopulations is likely to be driven by the availability of new biological niches, consistent with the hypothesis of punctuated equilibria. The hypothesis also has implications for the ongoing debate on the role of genetic drift in genome evolution.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

