Chemokine-driven lymphocyte infiltration: an early intratumoural event determining long-term survival in resectable hepatocellular carcinoma
- PMID: 21930732
- PMCID: PMC3273680
- DOI: 10.1136/gutjnl-2011-300509
Chemokine-driven lymphocyte infiltration: an early intratumoural event determining long-term survival in resectable hepatocellular carcinoma
Abstract
Objective: Hepatocellular carcinoma (HCC) is a heterogeneous disease with poor prognosis and limited methods for predicting patient survival. The nature of the immune cells that infiltrate tumours is known to impact clinical outcome. However, the molecular events that regulate this infiltration require further understanding. Here the ability of immune genes expressed in the tumour microenvironment to predict disease progression was investigated.
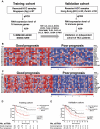
Methods: Using quantitative PCR, the expression of 14 immune genes in resected tumour tissues from 57 Singaporean patients was analysed. The nearest-template prediction method was used to derive and test a prognostic signature from this training cohort. The signature was then validated in an independent cohort of 98 patients from Hong Kong and Zurich. Intratumoural components expressing these critical immune genes were identified by in situ labelling. Regulation of these genes was analysed in vitro using the HCC cell line SNU-182.
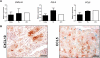
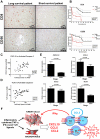
Results: The identified 14 immune-gene signature predicts patient survival in both the training cohort (p=0.0004 and HR=5.2) and the validation cohort (p=0.0051 and HR=2.5) irrespective of patient ethnicity and disease aetiology. Importantly, it predicts the survival of patients with early disease (stages I and II), for whom classical clinical parameters provide limited information. The lack of predictive power in late disease stages III and IV emphasises that a protective immune microenvironment has to be established early in order to impact disease progression significantly. This signature includes the chemokine genes CXCL10, CCL5 and CCL2, whose expression correlates with markers of T helper 1 (Th1), CD8(+) T and natural killer (NK) cells. Inflammatory cytokines (tumour necrosis factor α, interferon γ) and Toll-like receptor 3 ligands stimulate intratumoural production of these chemokines which drive tumour infiltration by T and NK cells, leading to enhanced cancer cell death.
Conclusion: A 14 immune-gene signature, which identifies molecular cues driving tumour infiltration by lymphocytes, accurately predicts survival of patients with HCC especially in early disease.
Conflict of interest statement
Figures



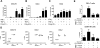
References
-
- El-Serag HB. Epidemiology of hepatocellular carcinoma in USA. Hepatol Res 2007;37(Suppl 2):S88–94 - PubMed
-
- Parkin DM, Bray F, Ferlay J, et al. Global cancer statistics, 2002. CA Cancer J Clin 2005;55:74–108 - PubMed
-
- Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet 2003;362:1907–17 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
