p63 regulates Satb1 to control tissue-specific chromatin remodeling during development of the epidermis
- PMID: 21930775
- PMCID: PMC3207288
- DOI: 10.1083/jcb.201101148
p63 regulates Satb1 to control tissue-specific chromatin remodeling during development of the epidermis
Abstract
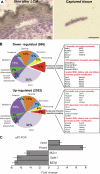
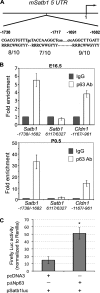
During development, multipotent progenitor cells establish tissue-specific programs of gene expression. In this paper, we show that p63 transcription factor, a master regulator of epidermal morphogenesis, executes its function in part by directly regulating expression of the genome organizer Satb1 in progenitor cells. p63 binds to a proximal regulatory region of the Satb1 gene, and p63 ablation results in marked reduction in the Satb1 expression levels in the epidermis. Satb1(-/-) mice show impaired epidermal morphology. In Satb1-null epidermis, chromatin architecture of the epidermal differentiation complex locus containing genes associated with epidermal differentiation is altered primarily at its central domain, where Satb1 binding was confirmed by chromatin immunoprecipitation-on-chip analysis. Furthermore, genes within this domain fail to be properly activated upon terminal differentiation. Satb1 expression in p63(+/-) skin explants treated with p63 small interfering ribonucleic acid partially restored the epidermal phenotype of p63-deficient mice. These data provide a novel mechanism by which Satb1, a direct downstream target of p63, contributes in epidermal morphogenesis via establishing tissue-specific chromatin organization and gene expression in epidermal progenitor cells.
Figures





References
-
- Agrelo R., Souabni A., Novatchkova M., Haslinger C., Leeb M., Komnenovic V., Kishimoto H., Gresh L., Kohwi-Shigematsu T., Kenner L., Wutz A. 2009. SATB1 defines the developmental context for gene silencing by Xist in lymphoma and embryonic cells. Dev. Cell. 16:507–516 10.1016/j.devcel.2009.03.006 - DOI - PMC - PubMed
-
- Antonini D., Rossi B., Han R., Minichiello A., Di Palma T., Corrado M., Banfi S., Zannini M., Brissette J.L., Missero C. 2006. An autoregulatory loop directs the tissue-specific expression of p63 through a long-range evolutionarily conserved enhancer. Mol. Cell. Biol. 26:3308–3318 10.1128/MCB.26.8.3308-3318.2006 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

