Discovery of the first insect nidovirus, a missing evolutionary link in the emergence of the largest RNA virus genomes
- PMID: 21931546
- PMCID: PMC3169540
- DOI: 10.1371/journal.ppat.1002215
Discovery of the first insect nidovirus, a missing evolutionary link in the emergence of the largest RNA virus genomes
Abstract
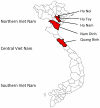
Nidoviruses with large genomes (26.3-31.7 kb; 'large nidoviruses'), including Coronaviridae and Roniviridae, are the most complex positive-sense single-stranded RNA (ssRNA+) viruses. Based on genome size, they are far separated from all other ssRNA+ viruses (below 19.6 kb), including the distantly related Arteriviridae (12.7-15.7 kb; 'small nidoviruses'). Exceptionally for ssRNA+ viruses, large nidoviruses encode a 3'-5'exoribonuclease (ExoN) that was implicated in controlling RNA replication fidelity. Its acquisition may have given rise to the ancestor of large nidoviruses, a hypothesis for which we here provide evolutionary support using comparative genomics involving the newly discovered first insect-borne nidovirus. This Nam Dinh virus (NDiV), named after a Vietnamese province, was isolated from mosquitoes and is yet to be linked to any pathology. The genome of this enveloped 60-80 nm virus is 20,192 nt and has a nidovirus-like polycistronic organization including two large, partially overlapping open reading frames (ORF) 1a and 1b followed by several smaller 3'-proximal ORFs. Peptide sequencing assigned three virion proteins to ORFs 2a, 2b, and 3, which are expressed from two 3'-coterminal subgenomic RNAs. The NDiV ORF1a/ORF1b frameshifting signal and various replicative proteins were tentatively mapped to canonical positions in the nidovirus genome. They include six nidovirus-wide conserved replicase domains, as well as the ExoN and 2'-O-methyltransferase that are specific to large nidoviruses. NDiV ORF1b also encodes a putative N7-methyltransferase, identified in a subset of large nidoviruses, but not the uridylate-specific endonuclease that - in deviation from the current paradigm - is present exclusively in the currently known vertebrate nidoviruses. Rooted phylogenetic inference by Bayesian and Maximum Likelihood methods indicates that NDiV clusters with roniviruses and that its branch diverged from large nidoviruses early after they split from small nidoviruses. Together these characteristics identify NDiV as the prototype of a new nidovirus family and a missing link in the transition from small to large nidoviruses.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Jenkins GM, Rambaut A, Pybus OG, Holmes EC. Rates of molecular evolution in RNA viruses: A quantitative phylogenetic analysis. J Mol Evol. 2002;54:156–165. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

