PCNA ubiquitination is important, but not essential for translesion DNA synthesis in mammalian cells
- PMID: 21931560
- PMCID: PMC3169526
- DOI: 10.1371/journal.pgen.1002262
PCNA ubiquitination is important, but not essential for translesion DNA synthesis in mammalian cells
Abstract
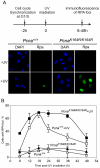
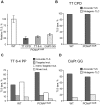
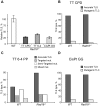
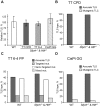
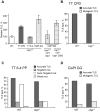
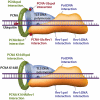
Translesion DNA synthesis (TLS) is a DNA damage tolerance mechanism in which specialized low-fidelity DNA polymerases bypass replication-blocking lesions, and it is usually associated with mutagenesis. In Saccharomyces cerevisiae a key event in TLS is the monoubiquitination of PCNA, which enables recruitment of the specialized polymerases to the damaged site through their ubiquitin-binding domain. In mammals, however, there is a debate on the requirement for ubiquitinated PCNA (PCNA-Ub) in TLS. We show that UV-induced Rpa foci, indicative of single-stranded DNA (ssDNA) regions caused by UV, accumulate faster and disappear more slowly in Pcna(K164R/K164R) cells, which are resistant to PCNA ubiquitination, compared to Pcna(+/+) cells, consistent with a TLS defect. Direct analysis of TLS in these cells, using gapped plasmids with site-specific lesions, showed that TLS is strongly reduced across UV lesions and the cisplatin-induced intrastrand GG crosslink. A similar effect was obtained in cells lacking Rad18, the E3 ubiquitin ligase which monoubiquitinates PCNA. Consistently, cells lacking Usp1, the enzyme that de-ubiquitinates PCNA exhibited increased TLS across a UV lesion and the cisplatin adduct. In contrast, cells lacking the Rad5-homologs Shprh and Hltf, which polyubiquitinate PCNA, exhibited normal TLS. Knocking down the expression of the TLS genes Rev3L, PolH, or Rev1 in Pcna(K164R/K164R) mouse embryo fibroblasts caused each an increased sensitivity to UV radiation, indicating the existence of TLS pathways that are independent of PCNA-Ub. Taken together these results indicate that PCNA-Ub is required for maximal TLS. However, TLS polymerases can be recruited to damaged DNA also in the absence of PCNA-Ub, and perform TLS, albeit at a significantly lower efficiency and altered mutagenic specificity.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
DNA-damage tolerance mediated by PCNA*Ub fusions in human cells is dependent on Rev1 but not Polη.Nucleic Acids Res. 2013 Aug;41(15):7356-69. doi: 10.1093/nar/gkt542. Epub 2013 Jun 12. Nucleic Acids Res. 2013. PMID: 23761444 Free PMC article.
-
HLTF and SHPRH are not essential for PCNA polyubiquitination, survival and somatic hypermutation: existence of an alternative E3 ligase.DNA Repair (Amst). 2011 Apr 3;10(4):438-44. doi: 10.1016/j.dnarep.2010.12.008. Epub 2011 Jan 26. DNA Repair (Amst). 2011. PMID: 21269891 Free PMC article.
-
Temporally distinct translesion synthesis pathways for ultraviolet light-induced photoproducts in the mammalian genome.DNA Repair (Amst). 2012 Jun 1;11(6):550-8. doi: 10.1016/j.dnarep.2012.03.007. Epub 2012 Apr 20. DNA Repair (Amst). 2012. PMID: 22521143
-
Regulation of DNA damage tolerance in mammalian cells by post-translational modifications of PCNA.Mutat Res. 2017 Oct;803-805:82-88. doi: 10.1016/j.mrfmmm.2017.06.004. Epub 2017 Jun 21. Mutat Res. 2017. PMID: 28666590 Review.
-
Ubiquitin-dependent regulation of translesion polymerases.Biochem Soc Trans. 2010 Feb;38(Pt 1):110-5. doi: 10.1042/BST0380110. Biochem Soc Trans. 2010. PMID: 20074045 Review.
Cited by
-
DNA-damage tolerance mediated by PCNA*Ub fusions in human cells is dependent on Rev1 but not Polη.Nucleic Acids Res. 2013 Aug;41(15):7356-69. doi: 10.1093/nar/gkt542. Epub 2013 Jun 12. Nucleic Acids Res. 2013. PMID: 23761444 Free PMC article.
-
Spartan/C1orf124 is important to prevent UV-induced mutagenesis.Cell Cycle. 2012 Sep 15;11(18):3395-402. doi: 10.4161/cc.21694. Epub 2012 Aug 16. Cell Cycle. 2012. PMID: 22894931 Free PMC article.
-
The auto-generated fragment of the Usp1 deubiquitylase is a physiological substrate of the N-end rule pathway.Mol Cell. 2012 Dec 28;48(6):926-33. doi: 10.1016/j.molcel.2012.10.012. Epub 2012 Nov 15. Mol Cell. 2012. PMID: 23159736 Free PMC article.
-
Different types of interaction between PCNA and PIP boxes contribute to distinct cellular functions of Y-family DNA polymerases.Nucleic Acids Res. 2015 Sep 18;43(16):7898-910. doi: 10.1093/nar/gkv712. Epub 2015 Jul 13. Nucleic Acids Res. 2015. PMID: 26170230 Free PMC article.
-
Requirement of Rad18 protein for replication through DNA lesions in mouse and human cells.Proc Natl Acad Sci U S A. 2012 May 15;109(20):7799-804. doi: 10.1073/pnas.1204105109. Epub 2012 Apr 30. Proc Natl Acad Sci U S A. 2012. PMID: 22547805 Free PMC article.
References
-
- Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem. 2005;74:317–353. - PubMed
-
- Livneh Z. Keeping mammalian mutation load in check. Regulation of the activity of error-prone DNA polymerases by p53 and p21. Cell Cycle. 2006;5:1918–1922. - PubMed
-
- Friedberg EC. Suffering in silence: The tolerance of DNA damage. Nature Rev Mol Cell Biol. 2005;6:943–953. - PubMed
-
- Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature. 2000;406:1015–1019. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

