Release of Ku and MRN from DNA ends by Mre11 nuclease activity and Ctp1 is required for homologous recombination repair of double-strand breaks
- PMID: 21931565
- PMCID: PMC3169521
- DOI: 10.1371/journal.pgen.1002271
Release of Ku and MRN from DNA ends by Mre11 nuclease activity and Ctp1 is required for homologous recombination repair of double-strand breaks
Abstract
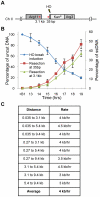
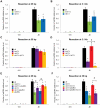
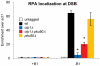
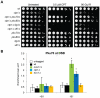
The multifunctional Mre11-Rad50-Nbs1 (MRN) protein complex recruits ATM/Tel1 checkpoint kinase and CtIP/Ctp1 homologous recombination (HR) repair factor to double-strand breaks (DSBs). HR repair commences with the 5'-to-3' resection of DNA ends, generating 3' single-strand DNA (ssDNA) overhangs that bind Replication Protein A (RPA) complex, followed by Rad51 recombinase. In Saccharomyces cerevisiae, the Mre11-Rad50-Xrs2 (MRX) complex is critical for DSB resection, although the enigmatic ssDNA endonuclease activity of Mre11 and the DNA-end processing factor Sae2 (CtIP/Ctp1 ortholog) are largely unnecessary unless the resection activities of Exo1 and Sgs1-Dna2 are also eliminated. Mre11 nuclease activity and Ctp1/CtIP are essential for DSB repair in Schizosaccharomyces pombe and mammals. To investigate DNA end resection in Schizo. pombe, we adapted an assay that directly measures ssDNA formation at a defined DSB. We found that Mre11 and Ctp1 are essential for the efficient initiation of resection, consistent with their equally crucial roles in DSB repair. Exo1 is largely responsible for extended resection up to 3.1 kb from a DSB, with an activity dependent on Rqh1 (Sgs1) DNA helicase having a minor role. Despite its critical function in DSB repair, Mre11 nuclease activity is not required for resection in fission yeast. However, Mre11 nuclease and Ctp1 are required to disassociate the MRN complex and the Ku70-Ku80 nonhomologous end-joining (NHEJ) complex from DSBs, which is required for efficient RPA localization. Eliminating Ku makes Mre11 nuclease activity dispensable for MRN disassociation and RPA localization, while improving repair of a one-ended DSB formed by replication fork collapse. From these data we propose that release of the MRN complex and Ku from DNA ends by Mre11 nuclease activity and Ctp1 is a critical step required to expose ssDNA for RPA localization and ensuing HR repair.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
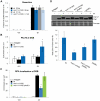
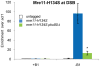
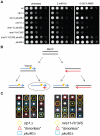

Similar articles
-
Nonhomologous End-Joining with Minimal Sequence Loss Is Promoted by the Mre11-Rad50-Nbs1-Ctp1 Complex in Schizosaccharomyces pombe.Genetics. 2017 May;206(1):481-496. doi: 10.1534/genetics.117.200972. Epub 2017 Mar 14. Genetics. 2017. PMID: 28292918 Free PMC article.
-
Ctp1-dependent clipping and resection of DNA double-strand breaks by Mre11 endonuclease complex are not genetically separable.Nucleic Acids Res. 2016 Sep 30;44(17):8241-9. doi: 10.1093/nar/gkw557. Epub 2016 Jun 20. Nucleic Acids Res. 2016. PMID: 27325741 Free PMC article.
-
Mre11 nuclease activity and Ctp1 regulate Chk1 activation by Rad3ATR and Tel1ATM checkpoint kinases at double-strand breaks.Mol Cell Biol. 2011 Feb;31(3):573-83. doi: 10.1128/MCB.00994-10. Epub 2010 Nov 22. Mol Cell Biol. 2011. PMID: 21098122 Free PMC article.
-
CtIP/Ctp1/Sae2, molecular form fit for function.DNA Repair (Amst). 2017 Aug;56:109-117. doi: 10.1016/j.dnarep.2017.06.013. Epub 2017 Jun 9. DNA Repair (Amst). 2017. PMID: 28623092 Free PMC article. Review.
-
A curious new role for MRN in Schizosaccharomyces pombe non-homologous end-joining.Curr Genet. 2018 Apr;64(2):359-364. doi: 10.1007/s00294-017-0760-1. Epub 2017 Oct 10. Curr Genet. 2018. PMID: 29018935 Free PMC article. Review.
Cited by
-
Part I-mechanism of adaptation: high nitric oxide adapted A549 cells show enhanced DNA damage response and activation of antiapoptotic pathways.Tumour Biol. 2014 Mar;35(3):2403-15. doi: 10.1007/s13277-013-1318-6. Epub 2013 Nov 16. Tumour Biol. 2014. PMID: 24241898
-
Increased sensitivity to ionizing radiation by targeting the homologous recombination pathway in glioma initiating cells.Mol Oncol. 2014 Dec;8(8):1603-15. doi: 10.1016/j.molonc.2014.06.012. Epub 2014 Jun 27. Mol Oncol. 2014. PMID: 25017126 Free PMC article.
-
Microhomology-Mediated End Joining: A Back-up Survival Mechanism or Dedicated Pathway?Trends Biochem Sci. 2015 Nov;40(11):701-714. doi: 10.1016/j.tibs.2015.08.006. Epub 2015 Oct 1. Trends Biochem Sci. 2015. PMID: 26439531 Free PMC article. Review.
-
MRE11A: a novel negative regulator of human DNA mismatch repair.Cell Mol Biol Lett. 2024 Mar 14;29(1):37. doi: 10.1186/s11658-024-00547-z. Cell Mol Biol Lett. 2024. PMID: 38486171 Free PMC article.
-
A Survey of Reported Disease-Related Mutations in the MRE11-RAD50-NBS1 Complex.Cells. 2020 Jul 13;9(7):1678. doi: 10.3390/cells9071678. Cells. 2020. PMID: 32668560 Free PMC article. Review.
References
-
- Aguilera A, Gomez-Gonzalez B. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet. 2008;9:204–217. - PubMed
-
- Pitcher RS, Wilson TE, Doherty AJ. New insights into NHEJ repair processes in prokaryotes. Cell Cycle. 2005;4:675–678. - PubMed
-
- Lees-Miller SP, Meek K. Repair of DNA double strand breaks by non-homologous end joining. Biochimie. 2003;85:1161–1173. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

