Integrated analysis of miRNA and mRNA expression in childhood medulloblastoma compared with neural stem cells
- PMID: 21931624
- PMCID: PMC3170291
- DOI: 10.1371/journal.pone.0023935
Integrated analysis of miRNA and mRNA expression in childhood medulloblastoma compared with neural stem cells
Abstract
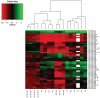
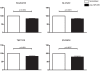
Medulloblastoma (MB) is the most common malignant brain tumor in children and a leading cause of cancer-related mortality and morbidity. Several molecular sub-types of MB have been identified, suggesting they may arise from distinct cells of origin. Data from animal models indicate that some MB sub-types arise from multipotent cerebellar neural stem cells (NSCs). Hence, microRNA (miRNA) expression profiles of primary MB samples were compared to CD133+ NSCs, aiming to identify deregulated miRNAs involved in MB pathogenesis. Expression profiling of 662 miRNAs in primary MB specimens, MB cell lines, and human CD133+ NSCs and CD133- neural progenitor cells was performed by qRT-PCR. Clustering analysis identified two distinct sub-types of MB primary specimens, reminiscent of sub-types obtained from their mRNA profiles. 21 significantly up-regulated and 12 significantly down-regulated miRNAs were identified in MB primary specimens relative to CD133+ NSCs (p<0.01). The majority of up-regulated miRNAs mapped to chromosomal regions 14q32 and 17q. Integration of the predicted targets of deregulated miRNAs with mRNA expression data from the same specimens revealed enrichment of pathways regulating neuronal migration, nervous system development and cell proliferation. Transient over-expression of a down-regulated miRNA, miR-935, resulted in significant down-regulation of three of the seven predicted miR-935 target genes at the mRNA level in a MB cell line, confirming the validity of this approach. This study represents the first integrated analysis of MB miRNA and mRNA expression profiles and is the first to compare MB miRNA expression profiles to those of CD133+ NSCs. We identified several differentially expressed miRNAs that potentially target networks of genes and signaling pathways that may be involved in the transformation of normal NSCs to brain tumor stem cells. Based on this integrative approach, our data provide an important platform for future investigations aimed at characterizing the role of specific miRNAs in MB pathogenesis.
Conflict of interest statement
Figures

Similar articles
-
Identification of suitable endogenous control genes for microRNA expression profiling of childhood medulloblastoma and human neural stem cells.BMC Res Notes. 2012 Sep 14;5:507. doi: 10.1186/1756-0500-5-507. BMC Res Notes. 2012. PMID: 22980291 Free PMC article.
-
Combined characterization of microRNA and mRNA profiles delineates early differentiation pathways of CD133+ and CD34+ hematopoietic stem and progenitor cells.Stem Cells. 2011 May;29(5):847-57. doi: 10.1002/stem.627. Stem Cells. 2011. PMID: 21394831 Free PMC article.
-
Sonic Hedgehog Medulloblastoma Cancer Stem Cells Mirnome and Transcriptome Highlight Novel Functional Networks.Int J Mol Sci. 2018 Aug 8;19(8):2326. doi: 10.3390/ijms19082326. Int J Mol Sci. 2018. PMID: 30096798 Free PMC article.
-
MicroRNAs expressed in neuronal differentiation and their associated pathways: Systematic review and bioinformatics analysis.Brain Res Bull. 2020 Apr;157:140-148. doi: 10.1016/j.brainresbull.2020.01.009. Epub 2020 Jan 13. Brain Res Bull. 2020. PMID: 31945407
-
From small to big: microRNAs as new players in medulloblastomas.Tumour Biol. 2013 Feb;34(1):9-15. doi: 10.1007/s13277-012-0579-9. Epub 2012 Nov 25. Tumour Biol. 2013. PMID: 23179395 Review.
Cited by
-
Aberrant methylation-mediated silencing of microRNAs contributes to HPV-induced anchorage independence.Oncotarget. 2016 Jul 12;7(28):43805-43819. doi: 10.18632/oncotarget.9698. Oncotarget. 2016. PMID: 27270309 Free PMC article.
-
SePIA: RNA and small RNA sequence processing, integration, and analysis.BioData Min. 2016 May 20;9:20. doi: 10.1186/s13040-016-0099-z. eCollection 2016. BioData Min. 2016. PMID: 27213017 Free PMC article.
-
The therapeutic and diagnostic potential of regulatory noncoding RNAs in medulloblastoma.Neurooncol Adv. 2019 May-Dec;1(1):vdz023. doi: 10.1093/noajnl/vdz023. Epub 2019 Sep 6. Neurooncol Adv. 2019. PMID: 31763623 Free PMC article. Review.
-
Downregulation of 14q32 microRNAs in Primary Human Desmoplastic Medulloblastoma.Front Oncol. 2013 Sep 25;3:254. doi: 10.3389/fonc.2013.00254. eCollection 2013. Front Oncol. 2013. PMID: 24093088 Free PMC article.
-
miRNome profiling in Duchenne muscular dystrophy; identification of asymptomatic and manifesting female carriers.Biosci Rep. 2021 Sep 30;41(9):BSR20211325. doi: 10.1042/BSR20211325. Biosci Rep. 2021. PMID: 34472584 Free PMC article.
References
-
- Giangaspero F, et al. Medulloblastoma, In: Louis DN, et al., editors. WHO Classification of Tumours of the Central Nervous System. International Agency for Research on Cancer (IARC): Lyon; 2007. pp. 132–140.
-
- Packer RJ, Rood BR, MacDonald TJ. Medulloblastoma: present concepts of stratification into risk groups. Pediatr Neurosurg. 2003;39(2):60–7. - PubMed
-
- Gottardo NG, Gajjar A. Current therapy for medulloblastoma. Curr Treat Options Neurol. 2006;8(4):319–34. - PubMed
-
- Crawford JR, MacDonald TJ, Packer RJ. Medulloblastoma in childhood: new biological advances. Lancet Neurol. 2007;6(12):1073–85. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

