Functional dissection of the TBK1 molecular network
- PMID: 21931631
- PMCID: PMC3169550
- DOI: 10.1371/journal.pone.0023971
Functional dissection of the TBK1 molecular network
Abstract
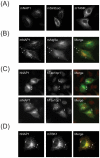
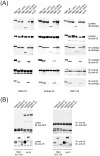
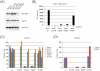
TANK-binding kinase 1 (TBK1) and inducible IκB-kinase (IKK-i) are central regulators of type-I interferon induction. They are associated with three adaptor proteins called TANK, Sintbad (or TBKBP1) and NAP1 (or TBKBP2, AZI2) whose functional relationship to TBK1 and IKK-i is poorly understood. We performed a systematic affinity purification-mass spectrometry approach to derive a comprehensive TBK1/IKK-i molecular network. The most salient feature of the network is the mutual exclusive interaction of the adaptors with the kinases, suggesting distinct alternative complexes. Immunofluorescence data indicated that the individual adaptors reside in different subcellular locations. TANK, Sintbad and NAP1 competed for binding of TBK1. The binding site for all three adaptors was mapped to the C-terminal coiled-coil 2 region of TBK1. Point mutants that affect binding of individual adaptors were used to reconstitute TBK1/IKK-i-deficient cells and dissect the functional relevance of the individual kinase-adaptor edges within the network. Using a microarray-derived gene expression signature of TBK1 in response virus infection or poly(I∶C) stimulation, we found that TBK1 activation was strictly dependent on the integrity of the TBK1/TANK interaction.
Conflict of interest statement
Figures





References
-
- Pichlmair A, Reis ESC. Innate Recognition of Viruses. Immunity. 2007;27:370–383. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

