Identification of mannose interacting residues using local composition
- PMID: 21931639
- PMCID: PMC3172211
- DOI: 10.1371/journal.pone.0024039
Identification of mannose interacting residues using local composition
Abstract
Background: Mannose binding proteins (MBPs) play a vital role in several biological functions such as defense mechanisms. These proteins bind to mannose on the surface of a wide range of pathogens and help in eliminating these pathogens from our body. Thus, it is important to identify mannose interacting residues (MIRs) in order to understand mechanism of recognition of pathogens by MBPs.
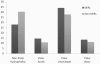
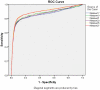
Results: This paper describes modules developed for predicting MIRs in a protein. Support vector machine (SVM) based models have been developed on 120 mannose binding protein chains, where no two chains have more than 25% sequence similarity. SVM models were developed on two types of datasets: 1) main dataset consists of 1029 mannose interacting and 1029 non-interacting residues, 2) realistic dataset consists of 1029 mannose interacting and 10320 non-interacting residues. In this study, firstly, we developed standard modules using binary and PSSM profile of patterns and got maximum MCC around 0.32. Secondly, we developed SVM modules using composition profile of patterns and achieved maximum MCC around 0.74 with accuracy 86.64% on main dataset. Thirdly, we developed a model on a realistic dataset and achieved maximum MCC of 0.62 with accuracy 93.08%. Based on this study, a standalone program and web server have been developed for predicting mannose interacting residues in proteins (http://www.imtech.res.in/raghava/premier/).
Conclusions: Compositional analysis of mannose interacting and non-interacting residues shows that certain types of residues are preferred in mannose interaction. It was also observed that residues around mannose interacting residues have a preference for certain types of residues. Composition of patterns/peptide/segment has been used for predicting MIRs and achieved reasonable high accuracy. It is possible that this novel strategy may be effective to predict other types of interacting residues. This study will be useful in annotating the function of protein as well as in understanding the role of mannose in the immune system.
Conflict of interest statement
Figures


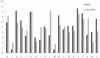
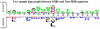
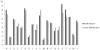
Similar articles
-
Prediction of GTP interacting residues, dipeptides and tripeptides in a protein from its evolutionary information.BMC Bioinformatics. 2010 Jun 3;11:301. doi: 10.1186/1471-2105-11-301. BMC Bioinformatics. 2010. PMID: 20525281 Free PMC article.
-
Identification of ATP binding residues of a protein from its primary sequence.BMC Bioinformatics. 2009 Dec 19;10:434. doi: 10.1186/1471-2105-10-434. BMC Bioinformatics. 2009. PMID: 20021687 Free PMC article.
-
SVM based prediction of RNA-binding proteins using binding residues and evolutionary information.J Mol Recognit. 2011 Mar-Apr;24(2):303-13. doi: 10.1002/jmr.1061. J Mol Recognit. 2011. PMID: 20677174
-
Identification of NAD interacting residues in proteins.BMC Bioinformatics. 2010 Mar 30;11:160. doi: 10.1186/1471-2105-11-160. BMC Bioinformatics. 2010. PMID: 20353553 Free PMC article.
-
Prediction of vitamin interacting residues in a vitamin binding protein using evolutionary information.BMC Bioinformatics. 2013 Feb 7;14:44. doi: 10.1186/1471-2105-14-44. BMC Bioinformatics. 2013. PMID: 23387468 Free PMC article.
Cited by
-
A hybrid approach for predicting transcription factors.Front Bioinform. 2024 Jul 25;4:1425419. doi: 10.3389/fbinf.2024.1425419. eCollection 2024. Front Bioinform. 2024. PMID: 39119181 Free PMC article.
-
NeuroPIpred: a tool to predict, design and scan insect neuropeptides.Sci Rep. 2019 Mar 26;9(1):5129. doi: 10.1038/s41598-019-41538-x. Sci Rep. 2019. PMID: 30914676 Free PMC article.
-
Community-based network study of protein-carbohydrate interactions in plant lectins using glycan array data.PLoS One. 2014 Apr 22;9(4):e95480. doi: 10.1371/journal.pone.0095480. eCollection 2014. PLoS One. 2014. PMID: 24755681 Free PMC article.
-
A web server for analysis, comparison and prediction of protein ligand binding sites.Biol Direct. 2016 Mar 25;11(1):14. doi: 10.1186/s13062-016-0118-5. Biol Direct. 2016. PMID: 27016210 Free PMC article.
-
Prediction of membrane transport proteins and their substrate specificities using primary sequence information.PLoS One. 2014 Jun 26;9(6):e100278. doi: 10.1371/journal.pone.0100278. eCollection 2014. PLoS One. 2014. PMID: 24968309 Free PMC article.
References
-
- Taroni C, Jones S, Thornton JM. Analysis and prediction of carbohydrate binding sites. Protein Eng. 2000;13(2):89–98. - PubMed
-
- Sompayrac L. Blackwell Science. Malden, MA: 1999. How the Immune System Works. pp. 17–19.
-
- Koch A, Melbye M, Sorensen P, Homoe P. Acute respiratory tract infections and mannose-binding lectin insufficiency during early childhood. JAMA. 2001;285:1316–1321. - PubMed
-
- Larsen F, Madsen HO, Sim RB, Koch C, Garred P. Disease-associated mutations in human mannose-binding lectin compromise oligomerization and activity of the final protein. J Biol Chem. 2004;279(20):21302–21311. - PubMed
-
- Hakomori S. Possible functions of tumor-associated carbohydrate antigens. Current Opinion in Immunology. 1991;1991;3(5):646–653. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

