In vivo RNAi screen reveals neddylation genes as novel regulators of Hedgehog signaling
- PMID: 21931660
- PMCID: PMC3169580
- DOI: 10.1371/journal.pone.0024168
In vivo RNAi screen reveals neddylation genes as novel regulators of Hedgehog signaling
Abstract
Hedgehog (Hh) signaling is highly conserved in all metazoan animals and plays critical roles in many developmental processes. Dysregulation of the Hh signaling cascade has been implicated in many diseases, including cancer. Although key components of the Hh pathway have been identified, significant gaps remain in our understanding of the regulation of individual Hh signaling molecules. Here, we report the identification of novel regulators of the Hh pathway, obtained from an in vivo RNA interference (RNAi) screen in Drosophila. By selectively targeting critical genes functioning in post-translational modification systems utilizing ubiquitin (Ub) and Ub-like proteins, we identify two novel genes (dUba3 and dUbc12) that negatively regulate Hh signaling activity. We provide in vivo and in vitro evidence illustrating that dUba3 and dUbc12 are essential components of the neddylation pathway; they function in an enzyme cascade to conjugate the ubiquitin-like NEDD8 modifier to Cullin proteins. Neddylation activates the Cullin-containing ubiquitin ligase complex, which in turn promotes the degradation of Cubitus interruptus (Ci), the downstream transcription factor of the Hh pathway. Our study reveals a conserved molecular mechanism of the neddylation pathway in Drosophila and sheds light on the complex post-translational regulations in Hh signaling.
Conflict of interest statement
Figures


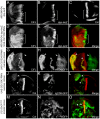

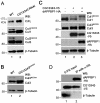
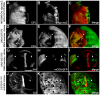

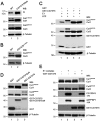
References
-
- Hooper JE, Scott MP. Communicating with Hedgehogs. Nat Rev Mol Cell Biol. 2005;6:306–317. - PubMed
-
- Varjosalo M, Taipale J. Hedgehog: functions and mechanisms. Genes Dev. 2008;22:2454–2472. - PubMed
-
- Noureddine MA, Donaldson TD, Thacker SA, Duronio RJ. Drosophila Roc1a encodes a RING-H2 protein with a unique function in processing the Hh signal transducer Ci by the SCF E3 ubiquitin ligase. Dev Cell. 2002;2:757–770. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

