Hepatic xenobiotic metabolizing enzyme and transporter gene expression through the life stages of the mouse
- PMID: 21931700
- PMCID: PMC3169610
- DOI: 10.1371/journal.pone.0024381
Hepatic xenobiotic metabolizing enzyme and transporter gene expression through the life stages of the mouse
Abstract
Background: Differences in responses to environmental chemicals and drugs between life stages are likely due in part to differences in the expression of xenobiotic metabolizing enzymes and transporters (XMETs). No comprehensive analysis of the mRNA expression of XMETs has been carried out through life stages in any species.
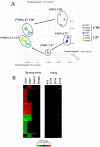
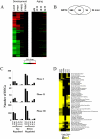
Results: Using full-genome arrays, the mRNA expression of all XMETs and their regulatory proteins was examined during fetal (gestation day (GD) 19), neonatal (postnatal day (PND) 7), prepubescent (PND32), middle age (12 months), and old age (18 and 24 months) in the C57BL/6J (C57) mouse liver and compared to adults. Fetal and neonatal life stages exhibited dramatic differences in XMET mRNA expression compared to the relatively minor effects of old age. The total number of XMET probe sets that differed from adults was 636, 500, 84, 5, 43, and 102 for GD19, PND7, PND32, 12 months, 18 months and 24 months, respectively. At all life stages except PND32, under-expressed genes outnumbered over-expressed genes. The altered XMETs included those in all of the major metabolic and transport phases including introduction of reactive or polar groups (Phase I), conjugation (Phase II) and excretion (Phase III). In the fetus and neonate, parallel increases in expression were noted in the dioxin receptor, Nrf2 components and their regulated genes while nuclear receptors and regulated genes were generally down-regulated. Suppression of male-specific XMETs was observed at early (GD19, PND7) and to a lesser extent, later life stages (18 and 24 months). A number of female-specific XMETs exhibited a spike in expression centered at PND7.
Conclusions: The analysis revealed dramatic differences in the expression of the XMETs, especially in the fetus and neonate that are partially dependent on gender-dependent factors. XMET expression can be used to predict life stage-specific responses to environmental chemicals and drugs.
Conflict of interest statement
Figures

References
-
- Francis GA, Fayard E, Picard F, Auwerx J. Nuclear receptors and the control of metabolism. Annu Rev Physiol. 2003;65:261–311. - PubMed
-
- Bleasby K, Castle JC, Roberts CJ, Cheng C, Bailey WJ, et al. Expression profiles of 50 xenobiotic transporter genes in humans and pre-clinical species: a resource for investigations into drug disposition. Xenobiotica. 2006;36:963–988. - PubMed
-
- Kohle C, Bock KW. Coordinate regulation of Phase I and II xenobiotic metabolisms by the Ah receptor and Nrf2. Biochem Pharmacol. 2007;73:1853–1862. - PubMed
-
- McGrath K, Palis J. Ontogeny of erythropoiesis in the mammalian embryo. Curr Top Dev Biol. 2008;82:1–22. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

