The priB gene of Klebsiella pneumoniae encodes a 104-amino acid protein that is similar in structure and function to Escherichia coli PriB
- PMID: 21931731
- PMCID: PMC3169591
- DOI: 10.1371/journal.pone.0024494
The priB gene of Klebsiella pneumoniae encodes a 104-amino acid protein that is similar in structure and function to Escherichia coli PriB
Abstract
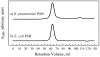
Primosome protein PriB is a single-stranded DNA-binding protein that serves as an accessory factor for PriA helicase-catalyzed origin-independent reinitiation of DNA replication in bacteria. A recent report describes the identification of a novel PriB protein in Klebsiella pneumoniae that is significantly shorter than most sequenced PriB homologs. The K. pneumoniae PriB protein is proposed to comprise 55 amino acid residues, in contrast to E. coli PriB which comprises 104 amino acid residues and has a length that is typical of most sequenced PriB homologs. Here, we report results of a sequence analysis that suggests that the priB gene of K. pneumoniae encodes a 104-amino acid PriB protein, akin to its E. coli counterpart. Furthermore, we have cloned the K. pneumoniae priB gene and purified the 104-amino acid K. pneumoniae PriB protein. Gel filtration experiments reveal that the K. pneumoniae PriB protein is a dimer, and equilibrium DNA binding experiments demonstrate that K. pneumoniae PriB's single-stranded DNA-binding activity is similar to that of E. coli PriB. These results indicate that the PriB homolog of K. pneumoniae is similar in structure and in function to that of E. coli.
Conflict of interest statement
Figures


References
-
- Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, et al. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. - PubMed
-
- Cox MM. The nonmutagenic repair of broken replication forks via recombination. Mutat Res. 2002;510:107–120. - PubMed
-
- McGlynn P, Lloyd RG. Recombinational repair and restart of damaged replication forks. Nat Rev Mol Cell Biol. 2002;3:859–870. - PubMed
-
- Heller RC, Marians KJ. Replisome assembly and the direct restart of stalled replication forks. Nat Rev Mol Cell Biol. 2006;7:932–943. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

