Protist-type lysozymes of the nematode Caenorhabditis elegans contribute to resistance against pathogenic Bacillus thuringiensis
- PMID: 21931778
- PMCID: PMC3169628
- DOI: 10.1371/journal.pone.0024619
Protist-type lysozymes of the nematode Caenorhabditis elegans contribute to resistance against pathogenic Bacillus thuringiensis
Abstract
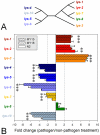
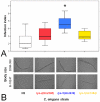
Pathogens represent a universal threat to other living organisms. Most organisms express antimicrobial proteins and peptides, such as lysozymes, as a protection against these challenges. The nematode Caenorhabditis elegans harbours 15 phylogenetically diverse lysozyme genes, belonging to two distinct types, the protist- or Entamoeba-type (lys genes) and the invertebrate-type (ilys genes) lysozymes. In the present study we characterized the role of several protist-type lysozyme genes in defence against a nematocidal strain of the Gram-positive bacterium Bacillus thuringiensis. Based on microarray and subsequent qRT-PCR gene expression analysis, we identified protist-type lysozyme genes as one of the differentially transcribed gene classes after infection. A functional genetic analysis was performed for three of these genes, each belonging to a distinct evolutionary lineage within the protist-type lysozymes (lys-2, lys-5, and lys-7). Their knock-out led to decreased pathogen resistance in all three cases, while an increase in resistance was observed when two out of three tested genes were overexpressed in transgenic lines (lys-5, lys-7, but not lys-2). We conclude that the lysozyme genes lys-5, lys-7, and possibly lys-2 contribute to resistance against B. thuringiensis, thus highlighting the particular role of lysozymes in the nematode's defence against pathogens.
Conflict of interest statement
Figures



References
-
- Beintema JJ, Terwisscha van Scheltinga AC. Plant lysozymes. In: Jollès P, editor. Lysozymes: Model enzymes in biochemistry and biology. Basel, Switzerland: Birkhäuser Verlag; 1996. pp. 75–86. - PubMed
-
- Fastrez J. Phage lysozymes. In: Jollès P, editor. Lysozymes: Model enzymes in biochemistry and biology. Basel, Switzerland: Birkhäuser Verlag; 1996. pp. 35–64.
-
- Höltje J-V. Bacterial lysozymes. In: Jollès P, editor. Lysozymes: Model enzymes in biochemistry and biology. Basel, Switzerland: Birkhäuser Verlag; 1996. pp. 65–74.
-
- Hultmark D. Insect lysozymes. In: Jollès P, editor. Lysozymes: Model enzymes in biochemistry and biology. Basel, Switzerland: Birkhäuser Verlag; 1996. pp. 87–101.
-
- Prager EM, Jollès P. Animal lysozymes c and g: An overview. In: Jollès P, editor. Lysozymes: Model enzymes in biochemistry and biology. Basel, Switzerland: Birkhäuser Verlag; 1996. pp. 9–31.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

