Expression of a novel antimicrobial peptide Penaeidin4-1 in creeping bentgrass (Agrostis stolonifera L.) enhances plant fungal disease resistance
- PMID: 21931807
- PMCID: PMC3171467
- DOI: 10.1371/journal.pone.0024677
Expression of a novel antimicrobial peptide Penaeidin4-1 in creeping bentgrass (Agrostis stolonifera L.) enhances plant fungal disease resistance
Abstract
Background: Turfgrass species are agriculturally and economically important perennial crops. Turfgrass species are highly susceptible to a wide range of fungal pathogens. Dollar spot and brown patch, two important diseases caused by fungal pathogens Sclerotinia homoecarpa and Rhizoctonia solani, respectively, are among the most severe turfgrass diseases. Currently, turf fungal disease control mainly relies on fungicide treatments, which raises many concerns for human health and the environment. Antimicrobial peptides found in various organisms play an important role in innate immune response.
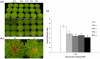
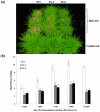
Methodology/principal findings: The antimicrobial peptide - Penaeidin4-1 (Pen4-1) from the shrimp, Litopenaeus setiferus has been reported to possess in vitro antifungal and antibacterial activities against various economically important fungal and bacterial pathogens. In this study, we have studied the feasibility of using this novel peptide for engineering enhanced disease resistance into creeping bentgrass plants (Agrostis stolonifera L., cv. Penn A-4). Two DNA constructs were prepared containing either the coding sequence of a single peptide, Pen4-1 or the DNA sequence coding for the transit signal peptide of the secreted tobacco AP24 protein translationally fused to the Pen4-1 coding sequence. A maize ubiquitin promoter was used in both constructs to drive gene expression. Transgenic turfgrass plants containing different DNA constructs were generated by Agrobacterium-mediated transformation and analyzed for transgene insertion and expression. In replicated in vitro and in vivo experiments under controlled environments, transgenic plants exhibited significantly enhanced resistance to dollar spot and brown patch, the two major fungal diseases in turfgrass. The targeting of Pen4-1 to endoplasmic reticulum by the transit peptide of AP24 protein did not significantly impact disease resistance in transgenic plants.
Conclusion/significance: Our results demonstrate the effectiveness of Pen4-1 in a perennial species against fungal pathogens and suggest a potential strategy for engineering broad-spectrum fungal disease resistance in crop species.
Conflict of interest statement
Figures





References
-
- Haydu J, Hodges A, Hall C. Economic impacts of the turfgrass and lawn care industry in the United States. University of Florida IFAS Extension website. 2006;23 Available: http://edis.ifas.ufl.edu/fe632. Accessed 2011 August.
-
- The national turfgrass research initiative. National Turfgrass Evaluation Program website. 2003;23 Available: http://www.ntep.org/pdf/turfinitiative.pdf. Accessed 2011 August.
-
- Chai B, Maqbool SB, Hajela RK, Green D, Vargas JM, et al. Cloning of a chitinase-like cDNA (hs2), its transfer to creeping bentgrass (Agrostis palustris Huds.) and development of brown patch (Rhizoctonia solani) disease resistant transgenic lines. Plant Sci. 2002;163:183–193.
-
- Guo Z, Bonos S, Meyer WA, Day PR, Belanger FC. Transgenic creeping bentgrass with delayed dollar spot symptoms. Mol Breed. 2003;11:95–101.
-
- Keymanesh K, Soltani S, Sardari S. Application of antimicrobial peptides in agriculture and food industry. World J Microbiol Biotechnol. 2009;25:933–944.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

