Genetic relationship between cocirculating Human enteroviruses species C
- PMID: 21931857
- PMCID: PMC3171481
- DOI: 10.1371/journal.pone.0024823
Genetic relationship between cocirculating Human enteroviruses species C
Abstract
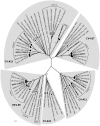
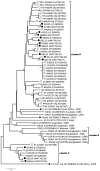
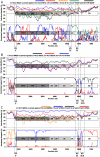
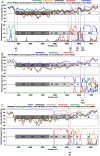
Recombination events between human enteroviruses (HEV) are known to occur frequently and to participate in the evolution of these viruses. In a previous study, we reported the isolation of a panel of viruses belonging to the Human enterovirus species C (HEV-C) that had been cocirculating in a small geographic area of Madagascar in 2002. This panel included type 2 vaccine-derived polioviruses (PV) that had caused several cases of acute flaccid paralysis in humans. Previous partial sequencing of the genome of these HEV-C isolates revealed considerable genetic diversity, mostly due to recombination. In the work presented herein, we carried out a more detailed characterization of the genomes of viruses from this collection. First, we determined the full VP1 sequence of 41 of these isolates of different types. These sequences were compared with those of HEV-C isolates obtained from other countries or in other contexts. The sequences of the Madagascan isolates of a given type formed specific clusters clearly differentiated from those formed by other strains of the same type isolated elsewhere. Second, we sequenced the entire genome of 10 viruses representing most of the lineages present in this panel. All but one of the genomes appeared to be mosaic assemblies of different genomic fragments generated by intra- and intertypic recombination. The location of the breakpoints suggested potential preferred genomic regions for recombination. Our results also suggest that recombination between type HEV-99 and other HEV-C may be quite rare. This first exhaustive genomic analysis of a panel of non-PV HEV-C cocirculating in a small human population highlights the high frequency of inter and intra-typic genetic recombination, constituting a widespread mechanism of genetic plasticity and continually shifting the HEV-C biodiversity.
Conflict of interest statement
Figures

References
-
- Racaniello VR. Picornaviridae: The Viruses and Their Replication. In: Knipe DM, editor. Fields Virology. Philadelphia: Wolters Kluwer Lippincott Williams & Wilkins; 2007. pp. 795–838. 5th ed.
-
- Picornaviridae study group website. Available: http://www.picornaviridae.1com/sequences/sequences.htm. Accessed 2011 Aug 24.
-
- International Committee on Taxonomy of Viruses website. . Available: http://www.ictvdb.org/. Accessed 2011 Aug 24.
-
- Lukashev AN. Role of recombination in evolution of enteroviruses. Rev Med Virol. 2005;15:157–167. - PubMed
-
- Lukashev AN. Recombination among picornaviruses. Rev Med Virol. 2010;20:327–337. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

