Hepatitis C virus proteins activate NRF2/ARE pathway by distinct ROS-dependent and independent mechanisms in HUH7 cells
- PMID: 21931870
- PMCID: PMC3172309
- DOI: 10.1371/journal.pone.0024957
Hepatitis C virus proteins activate NRF2/ARE pathway by distinct ROS-dependent and independent mechanisms in HUH7 cells
Abstract
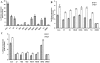
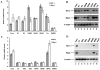
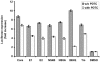
Hepatitis C virus (HCV) is a highly pathogenic human virus associated with liver fibrosis, steatosis, and cancer. In infected cells HCV induces oxidative stress. Here, we show that HCV proteins core, E1, E2, NS4B, and NS5A activate antioxidant defense Nrf2/ARE pathway via several independent mechanisms. This was demonstrated by the analysis of transient co-expression in Huh7 cells of HCV proteins and luciferase reporters. Expression, controlled by the promoters of stress-response genes or their minimal Nrf2-responsive elements, was studied using luminescence assay, RT-qPCR and/or Western-blot analysis. All five proteins induced Nrf2 activation by protein kinase C in response to accumulation of reactive oxygen species (ROS). In addition, expression of core, E1, E2, NS4B, and NS5A proteins resulted in the activation of Nrf2 in a ROS-independent manner. The effect of core and NS5A was mediated through casein kinase 2 and phosphoinositide-3 kinase, whereas those of NS4B, E1, and E2, were not mediated by either PKC, CK2, PI3K, p38, or ERK. Altogether, on the earliest stage of expression HCV proteins induced a strong up-regulation of the antioxidant defense system. These events may underlie the harmful effects of HCV-induced oxidative stress during acute stage of hepatitis C.
Conflict of interest statement
Figures
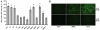
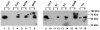
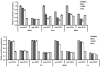
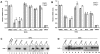
References
-
- Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis. 2005;5:558–567. - PubMed
-
- Nocente R, Ceccanti M, Bertazzoni G, Cammarota G, Silveri NG, et al. HCV infection and extrahepatic manifestations. Hepatogastroenterology. 2003;50:1149–1154. - PubMed
-
- Jacobson IM, Davis GL, El-Serag H, Negro F, Trepo C. Prevalence and challenges of liver diseases in patients with chronic hepatitis C virus infection. Clin Gastroenterol Hepatol. 2010;8:924–933; quiz e117. - PubMed
-
- National Institutes of Health. NIH Consensus Statement on Management of Hepatitis C: 2002. NIH Consens State Sci Statements. 2002;19:1–46. - PubMed
-
- Levrero M. Viral hepatitis and liver cancer: the case of hepatitis C. Oncogene. 2006;25:3834–3847. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

