Uridine phosphorylase from Trypanosoma cruzi: kinetic and chemical mechanisms
- PMID: 21932786
- PMCID: PMC3208269
- DOI: 10.1021/bi2013382
Uridine phosphorylase from Trypanosoma cruzi: kinetic and chemical mechanisms
Abstract
The reversible phosphorolysis of uridine to generate uracil and ribose 1-phosphate is catalyzed by uridine phosphorylase and is involved in the pyrimidine salvage pathway. We define the reaction mechanism of uridine phosphorylase from Trypanosoma cruzi by steady-state and pre-steady-state kinetics, pH-rate profiles, kinetic isotope effects from uridine, and solvent deuterium isotope effects. Initial rate and product inhibition patterns suggest a steady-state random kinetic mechanism. Pre-steady-state kinetics indicated no rate-limiting step after formation of the enzyme-products ternary complex, as no burst in product formation is observed. The limiting single-turnover rate constant equals the steady-state turnover number; thus, chemistry is partially or fully rate limiting. Kinetic isotope effects with [1'-(3)H]-, [1'-(14)C]-, and [5'-(14)C,1,3-(15)N(2)]uridine gave experimental values of (α-T)(V/K)(uridine) = 1.063, (14)(V/K)(uridine) = 1.069, and (15,β-15)(V/K)(uridine) = 1.018, in agreement with an A(N)D(N) (S(N)2) mechanism where chemistry contributes significantly to the overall rate-limiting step of the reaction. Density functional theory modeling of the reaction in gas phase supports an A(N)D(N) mechanism. Solvent deuterium kinetic isotope effects were unity, indicating that no kinetically significant proton transfer step is involved at the transition state. In this N-ribosyl transferase, proton transfer to neutralize the leaving group is not part of transition state formation, consistent with an enzyme-stabilized anionic uracil as the leaving group. Kinetic analysis as a function of pH indicates one protonated group essential for catalysis and for substrate binding.
Figures
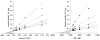








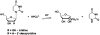



Similar articles
-
Constrained bonding environment in the Michaelis complex of Trypanosoma cruzi uridine phosphorylase.Biochemistry. 2012 Aug 28;51(34):6715-7. doi: 10.1021/bi300914q. Epub 2012 Aug 13. Biochemistry. 2012. PMID: 22870934 Free PMC article.
-
Transition-state analysis of Trypanosoma cruzi uridine phosphorylase-catalyzed arsenolysis of uridine.J Am Chem Soc. 2011 Jun 29;133(25):9923-31. doi: 10.1021/ja2031294. Epub 2011 Jun 3. J Am Chem Soc. 2011. PMID: 21599004 Free PMC article.
-
The kinetic mechanism of human uridine phosphorylase 1: Towards the development of enzyme inhibitors for cancer chemotherapy.Arch Biochem Biophys. 2010 May;497(1-2):35-42. doi: 10.1016/j.abb.2010.03.004. Epub 2010 Mar 11. Arch Biochem Biophys. 2010. PMID: 20226755
-
The crystal structure of Streptococcus pyogenes uridine phosphorylase reveals a distinct subfamily of nucleoside phosphorylases.Biochemistry. 2011 Aug 2;50(30):6549-58. doi: 10.1021/bi200707z. Epub 2011 Jul 8. Biochemistry. 2011. PMID: 21707079 Free PMC article.
-
Kinetic and chemical mechanisms of the fabG-encoded Streptococcus pneumoniae beta-ketoacyl-ACP reductase.Biochemistry. 2005 Dec 20;44(50):16753-65. doi: 10.1021/bi050947j. Biochemistry. 2005. PMID: 16342966
Cited by
-
Constrained bonding environment in the Michaelis complex of Trypanosoma cruzi uridine phosphorylase.Biochemistry. 2012 Aug 28;51(34):6715-7. doi: 10.1021/bi300914q. Epub 2012 Aug 13. Biochemistry. 2012. PMID: 22870934 Free PMC article.
-
Crystal Structure, Steady-State, and Pre-Steady-State Kinetics of Acinetobacter baumannii ATP Phosphoribosyltransferase.Biochemistry. 2024 Jan 16;63(2):230-240. doi: 10.1021/acs.biochem.3c00551. Epub 2023 Dec 27. Biochemistry. 2024. PMID: 38150593 Free PMC article.
-
The Evolution of Substrate Specificity by tRNA Modification Enzymes.Enzymes. 2017;41:51-88. doi: 10.1016/bs.enz.2017.03.002. Epub 2017 Apr 26. Enzymes. 2017. PMID: 28601226 Free PMC article. Review.
-
The Peculiar Case of the Hyper-thermostable Pyrimidine Nucleoside Phosphorylase from Thermus thermophilus*.Chembiochem. 2021 Apr 16;22(8):1385-1390. doi: 10.1002/cbic.202000679. Epub 2021 Jan 26. Chembiochem. 2021. PMID: 33258231 Free PMC article.
-
Pseudouridine: still mysterious, but never a fake (uridine)!RNA Biol. 2014;11(12):1540-54. doi: 10.4161/15476286.2014.992278. RNA Biol. 2014. PMID: 25616362 Free PMC article. Review.
References
-
- Paege LM, Schlenk F. Bacterial uracil riboside phosphorylase. Arch Biochem Biophys. 1952;40:42–49. - PubMed
-
- Pizzorno G, Cao D, Leffert JJ, Russell RL, Zhang D, Handschumacher RE. Homeostatic control of uridine and the role of uridine phosphorylase: a biological and clinical update. Biochim Biophys Acta. 2002;1587:133–144. - PubMed
-
- Cao D, Leffert JJ, McCabe J, Kim B, Pizzorno G. Abnormalities in uridine homeostatic regulation and pyrimidine nucleotide metabolism as a consequence of the deletion of the uridine phosphorylase gene. J Biol Chem. 2005;280:21169–21175. - PubMed
-
- Vita A, Huang CY, Magni G. Uridine phosphorylase from Escherichia coli B.: kinetic studies on the mechanism of catalysis. Arch Biochem Biophys. 1983;226:687–692. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

