Differential proteome analysis of human embryonic kidney cell line (HEK-293) following mycophenolic acid treatment
- PMID: 21933383
- PMCID: PMC3189873
- DOI: 10.1186/1477-5956-9-57
Differential proteome analysis of human embryonic kidney cell line (HEK-293) following mycophenolic acid treatment
Abstract
Background: Mycophenolic acid (MPA) is widely used as a post transplantation medicine to prevent acute organ rejection. In the present study we used proteomics approach to identify proteome alterations in human embryonic kidney cells (HEK-293) after treatment with therapeutic dose of MPA. Following 72 hours MPA treatment, total protein lysates were prepared, resolved by two dimensional gel electrophoresis and differentially expressed proteins were identified by QTOF-MS/MS analysis. Expressional regulations of selected proteins were further validated by real time PCR and Western blotting.
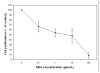
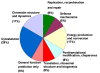
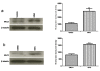
Results: The proliferation assay demonstrated that therapeutic MPA concentration causes a dose dependent inhibition of HEK-293 cell proliferation. A significant apoptosis was observed after MPA treatment, as revealed by caspase 3 activity. Proteome analysis showed a total of 12 protein spots exhibiting differential expression after incubation with MPA, of which 7 proteins (complement component 1 Q subcomponent-binding protein, electron transfer flavoprotein subunit beta, cytochrome b-c1 complex subunit, peroxiredoxin 1, thioredoxin domain-containing protein 12, myosin regulatory light chain 2, and profilin 1) showed significant increase in their expression. The expression of 5 proteins (protein SET, stathmin, 40S ribosomal protein S12, histone H2B type 1 A, and histone H2B type 1-C/E/F/G/I) were down-regulated. MPA mainly altered the proteins associated with the cytoskeleton (26%), chromatin structure/dynamics (17%) and energy production/conversion (17%). Both real time PCR and Western blotting confirmed the regulation of myosin regulatory light chain 2 and peroxiredoxin 1 by MPA treatment. Furthermore, HT-29 cells treated with MPA and total kidney cell lysate from MMF treated rats showed similar increased expression of myosin regulatory light chain 2.
Conclusion: The emerging use of MPA in diverse pathophysiological conditions demands in-depth studies to understand molecular basis of its therapeutic response. The present study identifies the myosin regulatory light chain 2 and peroxiredoxin 1 along with 10 other proteins showing significant regulation by MPA. Further characterization of these proteins may help to understand the diverse cellular effects of MPA in addition to its immunosuppressive activity.
Figures



References
-
- Nagai M, Natsumeda Y, Konno Y, Hoffman R, Irino S, Weber G. Selective up-regulation of type II inosine 5'-monophosphate dehydrogenase messenger RNA expression in human leukemias. Cancer Res. 1991;51:3886–3890. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

