Rhabdovirus accessory genes
- PMID: 21933691
- PMCID: PMC7114375
- DOI: 10.1016/j.virusres.2011.09.004
Rhabdovirus accessory genes
Abstract
The Rhabdoviridae is one of the most ecologically diverse families of RNA viruses with members infecting a wide range of organisms including placental mammals, marsupials, birds, reptiles, fish, insects and plants. The availability of complete nucleotide sequences for an increasing number of rhabdoviruses has revealed that their ecological diversity is reflected in the diversity and complexity of their genomes. The five canonical rhabdovirus structural protein genes (N, P, M, G and L) that are shared by all rhabdoviruses are overprinted, overlapped and interspersed with a multitude of novel and diverse accessory genes. Although not essential for replication in cell culture, several of these genes have been shown to have roles associated with pathogenesis and apoptosis in animals, and cell-to-cell movement in plants. Others appear to be secreted or have the characteristics of membrane-anchored glycoproteins or viroporins. However, most encode proteins of unknown function that are unrelated to any other known proteins. Understanding the roles of these accessory genes and the strategies by which rhabdoviruses use them to engage, divert and re-direct cellular processes will not only present opportunities to develop new anti-viral therapies but may also reveal aspects of cellar function that have broader significance in biology, agriculture and medicine.
Crown Copyright © 2011. Published by Elsevier B.V. All rights reserved.
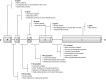
Figures





References
-
- Ahne W., Bjorklund H.V., Essbauer S., Fijan N., Kurath G., Winton J.R. Spring viremia of carp (SVC) Diseases of Aquatic Organisms. 2002;52:261–272. - PubMed
-
- Allison A.B., Palacios G., Travassos da Rosa A., Popov V.L., Lu L., Xiao S.Y., DeToy K., Briese T., Lipkin W.I., Keel M.K., Stallknecht D.E., Bishop G.R., Tesh R.B. Characterization of Durham virus, a novel rhabdovirus that encodes both a C and SH protein. Virus Research. 2011;155:112–122. - PMC - PubMed
-
- Ammayappan A., Kurath G., Thompson T., Vakharia V. A reverse genetics system for the Great Lakes strain of viral hemorrhagic septicemia virus: the NV gene is required for pathogenicity. Marine Biotechnology. 2010 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

