ChIP-Seq: technical considerations for obtaining high-quality data
- PMID: 21934668
- PMCID: PMC3541830
- DOI: 10.1038/ni.2117
ChIP-Seq: technical considerations for obtaining high-quality data
Abstract
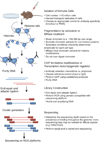
Chromatin immunoprecipitation followed by next-generation sequencing analysis (ChIP-Seq) is a powerful method with which to investigate the genome-wide distribution of chromatin-binding proteins and histone modifications in any genome with a known sequence. The application of this technique to a variety of developmental and differentiation systems has provided global views of the cis-regulatory elements, transcription factor function and epigenetic processes involved in the control of gene transcription. Here we describe several technical aspects of the ChIP-Seq assay that diminish bias and background noise and allow the consistent generation of high-quality data.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

