Integrating quantitative knowledge into a qualitative gene regulatory network
- PMID: 21935350
- PMCID: PMC3174175
- DOI: 10.1371/journal.pcbi.1002157
Integrating quantitative knowledge into a qualitative gene regulatory network
Abstract
Despite recent improvements in molecular techniques, biological knowledge remains incomplete. Any theorizing about living systems is therefore necessarily based on the use of heterogeneous and partial information. Much current research has focused successfully on the qualitative behaviors of macromolecular networks. Nonetheless, it is not capable of taking into account available quantitative information such as time-series protein concentration variations. The present work proposes a probabilistic modeling framework that integrates both kinds of information. Average case analysis methods are used in combination with Markov chains to link qualitative information about transcriptional regulations to quantitative information about protein concentrations. The approach is illustrated by modeling the carbon starvation response in Escherichia coli. It accurately predicts the quantitative time-series evolution of several protein concentrations using only knowledge of discrete gene interactions and a small number of quantitative observations on a single protein concentration. From this, the modeling technique also derives a ranking of interactions with respect to their importance during the experiment considered. Such a classification is confirmed by the literature. Therefore, our method is principally novel in that it allows (i) a hybrid model that integrates both qualitative discrete model and quantities to be built, even using a small amount of quantitative information, (ii) new quantitative predictions to be derived, (iii) the robustness and relevance of interactions with respect to phenotypic criteria to be precisely quantified, and (iv) the key features of the model to be extracted that can be used as a guidance to design future experiments.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
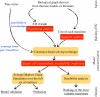




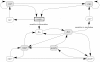
 ), either its increase (
), either its increase ( ) or its decrease (
) or its decrease ( ). Arrows between events depict the active effect of one event on another. Two transitions are absent when the system is subject to carbon starvation.
). Arrows between events depict the active effect of one event on another. Two transitions are absent when the system is subject to carbon starvation.
References
-
- Kauffman SA. Metabolic stability and epigenesis in randomly constructed genetic nets. J Theor Biol. 1969;22:437–467. - PubMed
-
- Thomas R. Regulatory networks seen as asynchronous automata: A logical description. J Theor Biol. 1991;153:1–23.
-
- Thomas R, Thieffry D, Kaufman M. Dynamical behaviour of biological regulatory networks–i. biological role of feedback loops and practical use of the concept of the loop-characteristic state. Bull Math Biol. 1995;57:247–76. - PubMed
-
- de Jong H. Modeling and simulation of genetic regulatory systems: a literature review. J Comput Biol. 2002;9:67–103. - PubMed
-
- Chen PCY, Chen JW. A Markovian approach to the control of genetic regulatory networks. Biosystems. 2007;90:535–545. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

