A phylometagenomic exploration of oceanic alphaproteobacteria reveals mitochondrial relatives unrelated to the SAR11 clade
- PMID: 21935411
- PMCID: PMC3173451
- DOI: 10.1371/journal.pone.0024457
A phylometagenomic exploration of oceanic alphaproteobacteria reveals mitochondrial relatives unrelated to the SAR11 clade
Abstract
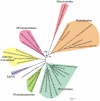
Background: According to the endosymbiont hypothesis, the mitochondrial system for aerobic respiration was derived from an ancestral Alphaproteobacterium. Phylogenetic studies indicate that the mitochondrial ancestor is most closely related to the Rickettsiales. Recently, it was suggested that Candidatus Pelagibacter ubique, a member of the SAR11 clade that is highly abundant in the oceans, is a sister taxon to the mitochondrial-Rickettsiales clade. The availability of ocean metagenome data substantially increases the sampling of Alphaproteobacteria inhabiting the oxygen-containing waters of the oceans that likely resemble the originating environment of mitochondria.
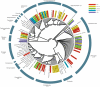
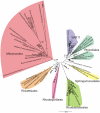
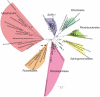
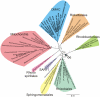
Methodology/principal findings: We present a phylogenetic study of the origin of mitochondria that incorporates metagenome data from the Global Ocean Sampling (GOS) expedition. We identify mitochondrially related sequences in the GOS dataset that represent a rare group of Alphaproteobacteria, designated OMAC (Oceanic Mitochondria Affiliated Clade) as the closest free-living relatives to mitochondria in the oceans. In addition, our analyses reject the hypothesis that the mitochondrial system for aerobic respiration is affiliated with that of the SAR11 clade.
Conclusions/significance: Our results allude to the existence of an alphaproteobacterial clade in the oxygen-rich surface waters of the oceans that represents the closest free-living relative to mitochondria identified thus far. In addition, our findings underscore the importance of expanding the taxonomic diversity in phylogenetic analyses beyond that represented by cultivated bacteria to study the origin of mitochondria.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

